Cell-specific spatial profiling of targeted protein expression to characterize the impact of intracortical microelectrode implantation on neuronal health
- PMID: 39479901
- PMCID: PMC11525954
- DOI: 10.1039/d4tb01628a
Cell-specific spatial profiling of targeted protein expression to characterize the impact of intracortical microelectrode implantation on neuronal health
Abstract
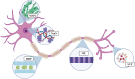
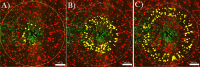
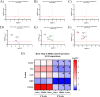
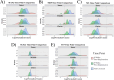
Intracortical microelectrode arrays (MEAs) can record neuronal activity and advance brain-computer interface (BCI) devices. Implantation of the invasive MEA kills local neurons, which has been documented using immunohistochemistry (IHC). Neuronal nuclear protein (NeuN), a protein that lines the nuclei of exclusively neuronal cells, has been used as a marker for neuronal health and survival for decades in neuroscience and neural engineering. NeuN staining is often used to describe the neuronal response to intracortical microelectrode array (MEA) implantation. However, IHC is semiquantitative, relying on intensity readings rather than directly counting expressed proteins. To supplement previous IHC studies, we evaluated the expression of proteins representing different aspects of neuronal structure or function: microtubule-associated protein 2 (MAP2), neurofilament light (NfL), synaptophysin (SYP), myelin basic protein (MBP), and oligodendrocyte transcription factor 2 (OLIG2) following a neural injury caused by intracortical MEA implantation. Together, these five proteins evaluate the cytoskeletal structure, neurotransmitter release, and myelination of neurons. To fully evaluate neuronal health in NeuN-positive (NeuN+) regions, we only quantified protein expression in NeuN+ regions, making this the first-ever cell-specific spatial profiling evaluation of targeted proteins by multiplex immunochemistry following MEA implantation. We performed our protein quantification along with NeuN IHC to compare the results of the two techniques directly. We found that NeuN immunohistochemical analysis does not show the same trends as MAP2, NfL, SYP, MBP, and OLIG2 expression. Further, we found that all five quantified proteins show a decreased expression pattern that aligns more with historic intracortical MEA recording performance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Badakva A. M. Miller N. V. Zobova L. N. [Artificial Feedback for Invasive Brain-Computer Interfaces] Fiziol. Chel. 2016;42(1):128–136. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

