The Circulating Proteome─Technological Developments, Current Challenges, and Future Trends
- PMID: 39479990
- PMCID: PMC11629384
- DOI: 10.1021/acs.jproteome.4c00586
The Circulating Proteome─Technological Developments, Current Challenges, and Future Trends
Abstract
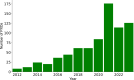
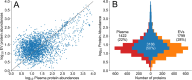
Recent improvements in proteomics technologies have fundamentally altered our capacities to characterize human biology. There is an ever-growing interest in using these novel methods for studying the circulating proteome, as blood offers an accessible window into human health. However, every methodological innovation and analytical progress calls for reassessing our existing approaches and routines to ensure that the new data will add value to the greater biomedical research community and avoid previous errors. As representatives of HUPO's Human Plasma Proteome Project (HPPP), we present our 2024 survey of the current progress in our community, including the latest build of the Human Plasma Proteome PeptideAtlas that now comprises 4608 proteins detected in 113 data sets. We then discuss the updates of established proteomics methods, emerging technologies, and investigations of proteoforms, protein networks, extracellualr vesicles, circulating antibodies and microsamples. Finally, we provide a prospective view of using the current and emerging proteomics tools in studies of circulating proteins.
Keywords: PTM; PeptideAtlas; affinity; biomarker discovery; blood; extracellular vesicle; mass spectrometry; microsampling; plasma; serum.
Conflict of interest statement
The authors declare the following competing financial interest(s): At the time of the first submission, D.H. was an employee of Bruker Scientific, K.K.P. was an employee of Freenome, and S.A. was an employee of Alkahest. The other authors declare that they have no conflict of interest.
Figures

References
-
- Omenn G. S.; et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 2005, 5, 3226–3245. 10.1002/pmic.200500358. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

