Adaptation to zinc restriction in Streptococcus agalactiae: role of the ribosomal protein and zinc-importers regulated by AdcR
- PMID: 39480081
- PMCID: PMC11580457
- DOI: 10.1128/msphere.00614-24
Adaptation to zinc restriction in Streptococcus agalactiae: role of the ribosomal protein and zinc-importers regulated by AdcR
Abstract
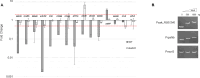
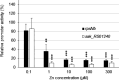
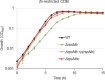
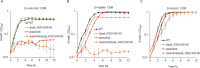
Zinc (Zn) is an essential cofactor for numerous bacterial proteins and altering Zn availability is an important component of host innate immunity. During infection, adaptation to both Zn deprivation and excess is critical for pathogenic bacteria development. To understand the adaptive responses to Zn availability of Streptococcus agalactiae, a pathogen causing invasive infections of neonates, global transcriptional profiling was conducted. Results highlight that in response to Zn limitation, genes belonging to the AdcR regulon, the master regulator of Zn homeostasis in streptococci, were overexpressed. Through a combination of in silico analysis and experimental validation, new AdcR-regulated targets were identified. Among them, we identified a duplicated ribosomal protein, RpsNb, and an ABC transporter, and examined the role of these genes in bacterial growth under Zn-restricted conditions. Our results indicated that, during Zn restriction, both the RpsNb protein and a potential secondary Zn transporter are important for S. agalactiae adaptation to Zn deficiency.
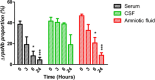
Importance: Streptococcus agalactiae is a bacterial human pathobiont causing invasive diseases in neonates. Upon infection, S. agalactiae is presented with Zn limitation and excess but the genetic systems that allow bacterial adaptation to these conditions remain largely undefined. A comprehensive analysis of S. agalactiae global transcriptional response to Zn availability shows that this pathogen manages Zn limitation mainly through upregulation of the AdcR regulon. We demonstrate that several AdcR-regulated genes are important for bacterial growth during Zn deficiency, including human biological fluids. Taken together, these findings reveal new mechanisms of S. agalactiae adaptation under conditions of metal deprivation.
Keywords: Streptococcus agalactiae; duplicated ribosomal proteins; gene regulation; metal ABC-transporter; zinc homeostasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
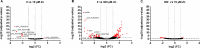



References
-
- Russell NJ, Seale AC, O’Driscoll M, O’Sullivan C, Bianchi-Jassir F, Gonzalez-Guarin J, Lawn JE, Baker CJ, Bartlett L, Cutland C, et al. . 2017. Maternal colonization with group B Streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin Infect Dis 65:S100–S111. doi:10.1093/cid/cix658 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
