Tripartite motif 25 inhibits protein aggregate degradation during PRRSV infection by suppressing p62-mediated autophagy
- PMID: 39480084
- PMCID: PMC11575163
- DOI: 10.1128/jvi.01437-24
Tripartite motif 25 inhibits protein aggregate degradation during PRRSV infection by suppressing p62-mediated autophagy
Abstract
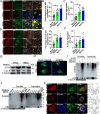
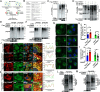
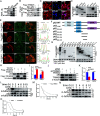
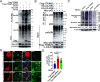
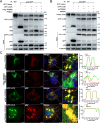
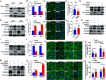
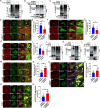
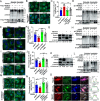
Viral infection causes endoplasmic reticulum stress and protein metabolism disorder, influencing protein aggregates formation or degradation that originate from misfolded proteins. The mechanism by which host proteins are involved in the above process remains largely unknown. The present study found that porcine reproductive and respiratory syndrome virus (PRRSV) infection promoted the degradation of intracellular ubiquitinated protein aggregates via activating autophagy. The host cell E3 ligase tripartite motif-containing (TRIM)25 promoted the recruitment and aggregation of polyubiquitinated proteins and impeded their degradation caused by PRRSV. TRIM25 interacted with ubiquitinated aggregates and was part of the aggregates complex. Next, the present study investigated the mechanisms by which TRIM25 inhibited the degradation of protein aggregates, and it was found that TRIM25 interacted with both Kelch-like ECH-associated protein 1 (KEAP1) and nuclear factor E2-related factor 2 (Nrf2), facilitated the nuclear translocation of Nrf2 by targeting KEAP1 for K48-linked ubiquitination and proteasome degradation, and activated Nrf2-mediated p62 expression. Further studies indicated that TRIM25 interacted with p62 and promoted its K63-linked ubiquitination via its E3 ligase activity and thus caused impairment of its oligomerization, aggregation, and recruitment for the autophagic protein LC3, leading to the suppression of autophagy activation. Besides, TRIM25 also suppressed the p62-mediated recruitment of ubiquitinated aggregates. Activation of autophagy decreased the accumulation of protein aggregates caused by TRIM25 overexpression, and inhibition of autophagy decreased the degradation of protein aggregates caused by TRIM25 knockdown. The current results also showed that TRIM25 inhibited PRRSV replication by inhibiting the KEAP1-Nrf2-p62 axis-mediated autophagy. Taken together, the present findings showed that the PRRSV replication restriction factor TRIM25 inhibited the degradation of ubiquitinated protein aggregates during viral infection by suppressing p62-mediated autophagy.IMPORTANCESequestration of protein aggregates and their subsequent degradation prevents proteostasis imbalance and cytotoxicity. The mechanisms controlling the turnover of protein aggregates during viral infection are mostly unknown. The present study found that porcine reproductive and respiratory syndrome virus (PRRSV) infection promoted the autophagic degradation of ubiquitinated protein aggregates, whereas tripartite motif-containing (TRIM)25 reversed this process. It was also found that TRIM25 promoted the expression of p62 by activating the Kelch-like ECH-associated protein 1 (KEAP1) and nuclear factor E2-related factor 2 (Nrf2) pathway and simultaneously prevented the oligomerization of p62 by promoting its K63-linked ubiquitination, thus suppressing its recruitment of the autophagic adaptor protein LC3 and ubiquitinated aggregates, leading to the inhibition of PRRSV-induced autophagy activation and the autophagic degradation of protein aggregates. The present study identified a new mechanism of protein aggregate turnover during viral infection and provided new insights for understanding the pathogenic mechanism of PRRSV.
Keywords: aggregates; autophagy; misfolded protein; porcine reproductive and respiratory syndrome virus; tripartite motif-containing 25.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

