Cyclophilin A facilitates HIV-1 integration
- PMID: 39480090
- PMCID: PMC11575316
- DOI: 10.1128/jvi.00947-24
Cyclophilin A facilitates HIV-1 integration
Abstract
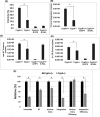
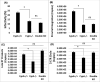
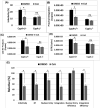
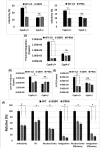
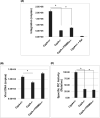
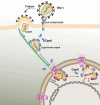
Cyclophilin A (CypA) binds to the HIV-1 capsid to facilitate reverse transcription and nuclear entry and counter the antiviral activity of TRIM5α. Interestingly, recent studies suggest that the capsid enters the nucleus of an infected cell and uncoats prior to integration. We have previously reported that the capsid protein regulates HIV-1 integration. Therefore, we probed whether CypA-capsid interaction also regulates this post-nuclear entry step. First, we challenged CypA-expressing (CypA+/+) and CypA-depleted (CypA-/-) cells with HIV-1 and quantified the levels of provirus. CypA-depletion significantly reduced integration, an effect that was independent of CypA's effect on reverse transcription, nuclear entry, and the presence or absence of TRIM5α. In addition, cyclosporin A, an inhibitor that disrupts CypA-capsid binding, inhibited proviral integration in CypA+/+ cells but not in CypA-/- cells. HIV-1 capsid mutants (G89V and P90A) deficient in CypA binding were also blocked at the integration step in CypA+/+ cells but not in CypA-/- cells. Then, to understand the mechanism, we assessed the integration activity of the HIV-1 preintegration complexes (PICs) extracted from acutely infected cells. PICs from CypA-/- cells retained lower integration activity in vitro compared to those from CypA+/+ cells. PICs from cells depleted of both CypA and TRIM5α also had lower activity, suggesting that CypA's effect on PIC was independent of TRIM5α. Finally, CypA protein specifically stimulated PIC activity, as this effect was significantly blocked by CsA. Collectively, these results provide strong evidence that CypA directly promotes HIV-1 integration, a previously unknown role of this host factor in the nucleus of an infected cell.
Importance: Interaction between the HIV-1 capsid and host cellular factors is essential for infection. However, the molecular details and functional consequences of viral-host factor interactions during HIV-1 infection are not fully understood. Over 30 years ago, Cyclophilin A (CypA) was identified as the first host protein to bind to the HIV-1 capsid. Now it is established that CypA-capsid interaction promotes reverse transcription and nuclear entry of HIV-1. In addition, CypA blocks TRIM5α-mediated restriction of HIV-1. In this report, we show that CypA promotes the post-nuclear entry step of HIV-1 integration by binding to the viral capsid. Notably, we show that CypA stimulates the viral DNA integration activity of the HIV-1 preintegration complex. Collectively, our studies identify a novel role of CypA during the early steps of HIV-1 infection. This new knowledge is important because recent reports suggest that an operationally intact HIV-1 capsid enters the nucleus of an infected cell.
Keywords: Cyclophilin A (CypA); capsid; human immunodeficiency virus (HIV); integration; reverse transcription nuclear entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
Cyclophilin A Facilitates HIV-1 DNA Integration.bioRxiv [Preprint]. 2024 Jun 17:2024.06.15.599180. doi: 10.1101/2024.06.15.599180. bioRxiv. 2024. Update in: J Virol. 2024 Nov 19;98(11):e0094724. doi: 10.1128/jvi.00947-24. PMID: 38948800 Free PMC article. Updated. Preprint.
References
-
- Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. 1999. Both memory and CD45RA+/CD62L+ naive CD4(+) T cells are infected in human immunodeficiency virus type 1-infected individuals. J Virol 73:6430–6435. doi:10.1128/JVI.73.8.6430-6435.1999 - DOI - PMC - PubMed
-
- Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi:10.1128/jvi.78.3.1160-1168.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

