Breast cancers that disseminate to bone marrow acquire aggressive phenotypes through CX43-related tumor-stroma tunnels
- PMID: 39480488
- PMCID: PMC11645149
- DOI: 10.1172/JCI170953
Breast cancers that disseminate to bone marrow acquire aggressive phenotypes through CX43-related tumor-stroma tunnels
Abstract
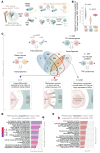
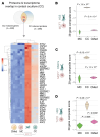
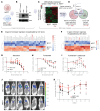
Estrogen receptor-positive (ER+) breast cancer commonly disseminates to bone marrow, where interactions with mesenchymal stromal cells (MSCs) shape disease trajectory. We modeled these interactions with tumor-MSC co-cultures and used an integrated transcriptome-proteome-network-analyses workflow to identify a comprehensive catalog of contact-induced changes. Conditioned media from MSCs failed to recapitulate genes and proteins, some borrowed and others tumor-intrinsic, induced in cancer cells by direct contact. Protein-protein interaction networks revealed the rich connectome between "borrowed" and "intrinsic" components. Bioinformatics prioritized one of the borrowed components, CCDC88A/GIV, a multi-modular metastasis-related protein that has recently been implicated in driving a hallmark of cancer, growth signaling autonomy. MSCs transferred GIV protein to ER+ breast cancer cells (that lack GIV) through tunnelling nanotubes via connexin (Cx)43-facilitated intercellular transport. Reinstating GIV alone in GIV-negative breast cancer cells reproduced approximately 20% of both the borrowed and the intrinsic gene induction patterns from contact co-cultures; conferred resistance to anti-estrogen drugs; and enhanced tumor dissemination. Findings provide a multiomic insight into MSC→tumor cell intercellular transport and validate how transport of one such candidate, GIV, from the haves (MSCs) to have-nots (ER+ breast cancer) orchestrates aggressive disease states.
Keywords: Bioinformatics; Bone marrow; Breast cancer; Oncology.
Conflict of interest statement
Figures






Update of
-
Breast Cancers That Disseminate to Bone Marrow Acquire Aggressive Phenotypes through CX43-related Tumor-Stroma Tunnels.bioRxiv [Preprint]. 2024 Aug 10:2023.03.18.533175. doi: 10.1101/2023.03.18.533175. bioRxiv. 2024. Update in: J Clin Invest. 2024 Oct 31;134(24):e170953. doi: 10.1172/JCI170953. PMID: 36993616 Free PMC article. Updated. Preprint.

