TMED4 facilitates regulatory T cell suppressive function via ROS homeostasis in tumor and autoimmune mouse models
- PMID: 39480507
- PMCID: PMC11684806
- DOI: 10.1172/JCI179874
TMED4 facilitates regulatory T cell suppressive function via ROS homeostasis in tumor and autoimmune mouse models
Abstract
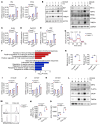
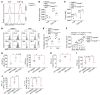
Endoplasmic reticulum stress (ERS) plays crucial roles in maintaining Treg stability and function, yet the underlying mechanism remains largely unexplored. Here, we demonstrate that (Tmed4ΔTreg) mice with Treg-specific KO of ERS-related protein transmembrane p24 trafficking protein 4 (TMED4) had more Tregs with impaired Foxp3 stability, Treg signatures, and suppressive activity, which led to T cell hyperactivation and an exacerbated inflammatory phenotype and boosted antitumor immunity in mice. Mechanistically, loss of Tmed4 caused defects in ERS and a nuclear factor erythroid 2-related factor 2-related (NRF2-related) antioxidant response, which resulted in excessive ROS that reduced the Foxp3 stability and suppressive function of Tregs in an IRE1α/XBP1 axis-dependent manner. The abnormalities could be effectively rescued by the ROS scavenger, NRF2 inducer, or by forcible expression of IRE1α. Moreover, TMED4 suppressed IRE1α proteosome degradation via the ER-associated degradation (ERAD) system including the ER chaperone binding immunoglobulin protein (BIP). Our study reveals that TMED4 maintained the stability of Tregs and their suppressive function through IRE1α-dependent ROS and the NRF2-related antioxidant response.
Keywords: Adaptive immunity; Autoimmune diseases; Immunology; T cells.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

