Large-Scale Pharmacogenomics Analysis of Patients With Cancer Within the 100,000 Genomes Project Combining Whole-Genome Sequencing and Medical Records to Inform Clinical Practice
- PMID: 39481076
- PMCID: PMC11825504
- DOI: 10.1200/JCO.23.02761
Large-Scale Pharmacogenomics Analysis of Patients With Cancer Within the 100,000 Genomes Project Combining Whole-Genome Sequencing and Medical Records to Inform Clinical Practice
Abstract
Purpose: As part of the 100,000 Genomes Project, we set out to assess the potential viability and clinical impact of reporting genetic variants associated with drug-induced toxicity for patients with cancer recruited for whole-genome sequencing (WGS) as part of a genomic medicine service.
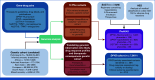
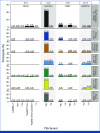
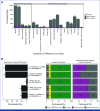
Methods: Germline WGS from 76,805 participants was analyzed for pharmacogenetic (PGx) variants in four genes (DPYD, NUDT15, TPMT, UGT1A1) associated with toxicity induced by five drugs used in cancer treatment (capecitabine, fluorouracil, mercaptopurine, thioguanine, irinotecan). Linking genomic data with prescribing and hospital incidence records, a phenome-wide association study (PheWAS) was performed to identify whether phenotypes indicative of adverse drug reactions (ADRs) were enriched in drug-exposed individuals with the relevant PGx variants. In a subset of 7,081 patients with cancer, DPYD variants were reported back to clinicians and outcomes were collected.
Results: We identified clinically relevant PGx variants across the four genes in 62.7% of participants in our cohort. Extending this to annual prescription numbers in England for the drugs affected by these PGx variants, approximately 14,540 patients per year could potentially benefit from a reduced dose or alternative drug to reduce the risk of ADRs. Validating PGx associations in a real-world data set, we found a significant association between PGx variants in DPYD and toxicity-related phenotypes in patients treated with capecitabine or fluorouracil. Reported DPYD variants were deemed informative for clinical decision making in a majority of cases.
Conclusion: Reporting PGx variants from germline WGS relevant to patients with cancer alongside primary findings related to their cancer can be clinically informative, informing prescribing to reduce the risk of ADRs. Extending the range of actionable variants to those found in patients of non-European ancestry is important and will extend the potential clinical impact.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Figures

References
-
- Meta-Analysis Group In Cancer. Lévy E, Piedbois P, et al. Toxicity of fluorouracil in patients with advanced colorectal cancer: Effect of administration schedule and prognostic factors. J Clin Oncol. 1998;16:3537–3541. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

