3D genome topology distinguishes molecular subgroups of medulloblastoma
- PMID: 39481374
- PMCID: PMC11639096
- DOI: 10.1016/j.ajhg.2024.10.003
3D genome topology distinguishes molecular subgroups of medulloblastoma
Abstract
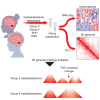
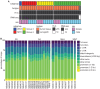
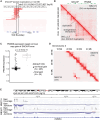
Four main medulloblastoma (MB) molecular subtypes have been identified based on transcriptional, DNA methylation, and genetic profiles. However, it is currently not known whether 3D genome architecture differs between MB subtypes. To address this question, we performed in situ Hi-C to reconstruct the 3D genome architecture of MB subtypes. In total, we generated Hi-C and matching transcriptome data for 28 surgical specimens and Hi-C data for one patient-derived xenograft. The average resolution of the Hi-C maps was 6,833 bp. Using these data, we found that insulation scores of topologically associating domains (TADs) were effective at distinguishing MB molecular subgroups. TAD insulation score differences between subtypes were globally not associated with differential gene expression, although we identified few exceptions near genes expressed in the lineages of origin of specific MB subtypes. Our study therefore supports the notion that TAD insulation scores can distinguish MB subtypes independently of their transcriptional differences.
Keywords: 3D genome; CNS tumor; Hi-C; cancer; medulloblastoma; transcriptome.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Northcott P.A., Korshunov A., Witt H., Hielscher T., Eberhart C.G., Mack S., Bouffet E., Clifford S.C., Hawkins C.E., French P., et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. - DOI - PMC - PubMed
-
- Hendrikse L.D., Haldipur P., Saulnier O., Millman J., Sjoboen A.H., Erickson A.W., Ong W., Gordon V., Coudière-Morrison L., Mercier A.L., et al. Failure of human rhombic lip differentiation underlies medulloblastoma formation. Nature. 2022;609:1021–1028. doi: 10.1038/s41586-022-05215-w. - DOI - PMC - PubMed
-
- Kool M., Korshunov A., Remke M., Jones D.T.W., Schlanstein M., Northcott P.A., Cho Y.-J., Koster J., Schouten-van Meeteren A., van Vuurden D., et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

