Senescence suppresses the integrated stress response and activates a stress-remodeled secretory phenotype
- PMID: 39481386
- PMCID: PMC11585442
- DOI: 10.1016/j.molcel.2024.10.003
Senescence suppresses the integrated stress response and activates a stress-remodeled secretory phenotype
Abstract
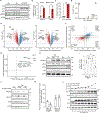
Senescence is a state of indefinite cell-cycle arrest associated with aging, cancer, and age-related diseases. Here, we find that translational deregulation, together with a corresponding maladaptive integrated stress response (ISR), is a hallmark of senescence that desensitizes senescent cells to stress. We present evidence that senescent cells maintain high levels of eIF2α phosphorylation, typical of ISR activation, but translationally repress production of the stress response activating transcription factor 4 (ATF4) by ineffective bypass of the inhibitory upstream open reading frames (uORFs). Surprisingly, ATF4 translation remains inhibited even after acute proteotoxic and amino acid starvation stressors, resulting in a highly diminished stress response. We also find that stress augments the senescence-associated secretory phenotype with sustained remodeling of inflammatory factors expression that is suppressed by non-uORF carrying ATF4 mRNA expression. Our results thus show that senescent cells possess a unique response to stress, which entails an increase in their inflammatory profile.
Keywords: ATF4; ER stress; ISR; SASP; integrated stress response; nanopore direct RNA sequencing; proteomics; ribosome sequencing; senescence; senescence-associated secretory phenotype; translation.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Senescence suppresses the integrated stress response and activates a stress-enhanced secretory phenotype.bioRxiv [Preprint]. 2023 Nov 18:2023.04.12.536613. doi: 10.1101/2023.04.12.536613. bioRxiv. 2023. Update in: Mol Cell. 2024 Nov 21;84(22):4454-4469.e7. doi: 10.1016/j.molcel.2024.10.003. PMID: 37609272 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

