Multi-omics after O-GlcNAc alteration identified cellular processes promoting aneuploidy after loss of O-GlcNAc transferase
- PMID: 39481848
- PMCID: PMC11585826
- DOI: 10.1016/j.molmet.2024.102060
Multi-omics after O-GlcNAc alteration identified cellular processes promoting aneuploidy after loss of O-GlcNAc transferase
Abstract
Objective: Pharmacologic or genetic manipulation of O-GlcNAcylation, an intracellular, single sugar post-translational modification, are difficult to interpret due to the pleotropic nature of O-GlcNAc and the vast signaling pathways it regulates.
Method: To address the pleotropic nature of O-GlcNAc, we employed either OGT (O-GlcNAc transferase), OGA (O-GlcNAcase) liver knockouts, or pharmacological inhibition of OGA coupled with multi-Omics analysis and bioinformatics.
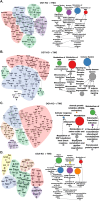
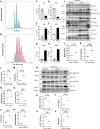
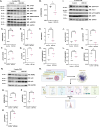
Results: We identified numerous genes, proteins, phospho-proteins, or metabolites that were either inversely or equivalently changed between conditions. Moreover, we identified pathways in OGT knockout samples associated with increased aneuploidy. To test and validate these pathways, we induced liver growth in OGT knockouts by partial hepatectomy. OGT knockout livers showed a robust aneuploidy phenotype with disruptions in mitosis, nutrient sensing, protein metabolism/amino acid metabolism, stress response, and HIPPO signaling demonstrating how OGT is essential in controlling aneuploidy pathways.
Conclusion: These data show how a multi-Omics platform can disentangle the pleotropic nature of O-GlcNAc to discern how OGT fine-tunes multiple cellular pathways involved in aneuploidy.
Keywords: Aneuploidy; Aurora kinase B; Cell cycle; O-GlcNAc; OGA; OGT; mTOR.
Copyright © 2024 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Chad Slawson reports financial support was provided by National Institutes of Health. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Update of
-
Multi-Omics after O-GlcNAc Alteration Identifies Cellular Processes Working Synergistically to Promote Aneuploidy.bioRxiv [Preprint]. 2024 Apr 16:2024.04.16.589379. doi: 10.1101/2024.04.16.589379. bioRxiv. 2024. Update in: Mol Metab. 2024 Dec;90:102060. doi: 10.1016/j.molmet.2024.102060. PMID: 38659829 Free PMC article. Updated. Preprint.
References
-
- Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

