The dual life of disordered lysine-rich domains of snoRNPs in rRNA modification and nucleolar compaction
- PMID: 39482307
- PMCID: PMC11528048
- DOI: 10.1038/s41467-024-53805-1
The dual life of disordered lysine-rich domains of snoRNPs in rRNA modification and nucleolar compaction
Abstract
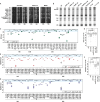
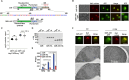
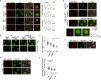
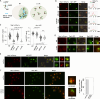
Intrinsically disordered regions (IDRs) are highly enriched in the nucleolar proteome but their physiological role in ribosome assembly remains poorly understood. Our study reveals the functional plasticity of the extremely abundant lysine-rich IDRs of small nucleolar ribonucleoprotein particles (snoRNPs) from protists to mammalian cells. We show in Saccharomyces cerevisiae that the electrostatic properties of this lysine-rich IDR, the KKE/D domain, promote snoRNP accumulation in the vicinity of nascent rRNAs, facilitating their modification. Under stress conditions reducing the rate of ribosome assembly, they are essential for nucleolar compaction and sequestration of key early-acting ribosome biogenesis factors, including RNA polymerase I, owing to their self-interaction capacity in a latent, non-rRNA-associated state. We propose that such functional plasticity of these lysine-rich IDRs may represent an ancestral eukaryotic regulatory mechanism, explaining how nucleolar morphology is continuously adapted to rRNA production levels.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

