Cohesin complex oligomerization maintains end-tethering at DNA double-strand breaks
- PMID: 39482358
- PMCID: PMC11735392
- DOI: 10.1038/s41556-024-01552-2
Cohesin complex oligomerization maintains end-tethering at DNA double-strand breaks
Abstract
DNA double-strand breaks (DSBs) must be repaired to ensure genome stability. Crucially, DSB-ends must be kept together for timely repair. In Saccharomyces cerevisiae, two pathways mediate DSB end-tethering. One employs the Mre11-Rad50-Xrs2 (MRX) complex to physically bridge DSB-ends. Another requires the conversion of DSB-ends into single-strand DNA (ssDNA) by Exo1, but the bridging proteins are unknown. We uncover that cohesin, its loader and Smc5/6 act with Exo1 to tether DSB-ends. Remarkably, cohesin specifically impaired in oligomerization fails to tether DSB-ends, revealing a function for cohesin oligomerization. In addition to the known importance of sister chromatid cohesion, microscopy-based microfluidic experiments unveil a role for cohesin in repair by ensuring DSB end-tethering. Altogether, our findings demonstrate that oligomerization of cohesin prevents DSB end-separation and promotes DSB repair, revealing a previously undescribed mode of action and role for cohesin in safeguarding genome integrity.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
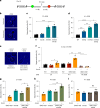

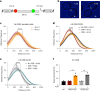
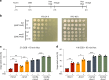

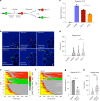
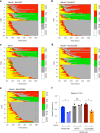



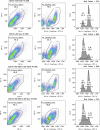

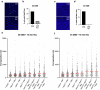
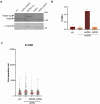
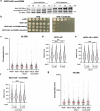
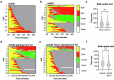
References
-
- Lobachev, K., Vitriol, E., Stemple, J., Resnick, M. A. & Bloom, K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol.14, 2107–2112 (2004). - PubMed
-
- Lee, K., Zhang, Y. & Lee, S. E. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature454, 543–546 (2008). - PubMed
MeSH terms
Substances
Grants and funding
- 281287/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- PJA-20191209432/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

