Proteome-scale recombinant standards and a robust high-speed search engine to advance cross-linking MS-based interactomics
- PMID: 39482464
- PMCID: PMC11621016
- DOI: 10.1038/s41592-024-02478-1
Proteome-scale recombinant standards and a robust high-speed search engine to advance cross-linking MS-based interactomics
Abstract
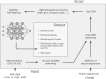
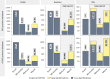
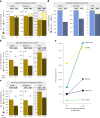
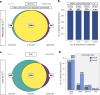
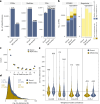
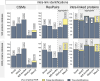
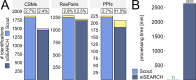
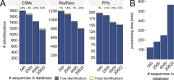
Advancing data analysis tools for proteome-wide cross-linking mass spectrometry (XL-MS) requires ground-truth standards that mimic biological complexity. Here we develop well-controlled XL-MS standards comprising hundreds of recombinant proteins that are systematically mixed for cross-linking. We use one standard dataset to guide the development of Scout, a search engine for XL-MS with MS-cleavable cross-linkers. Using other, independent standard datasets and published datasets, we benchmark the performance of Scout and existing XL-MS software. We find that Scout offers an excellent combination of speed, sensitivity and false discovery rate control. The results illustrate how our large recombinant standard can support the development of XL-MS analysis tools and evaluation of XL-MS results.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: F.L. is a shareholder and advisory board member of Absea Biotechnology Ltd and VantAI. T.C. is the co-founder of Absea Biotechnology Ltd. S.W. is an employee of Absea Biotechnology Ltd. The remaining authors declare no competing interests.
Figures


References
-
- Graziadei, A. & Rappsilber, J. Leveraging crosslinking mass spectrometry in structural and cell biology. Structure30, 37–54 (2022). - PubMed
-
- Liu, F., Rijkers, D. T., Post, H. & Heck, A. J. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods12, 1179–1184 (2015). - PubMed
-
- Lima, D. B. et al. SIM-XL: a powerful and user-friendly tool for peptide cross-linking analysis. J. Proteom.129, 51–55 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

