The 24-hour molecular landscape after exercise in humans reveals MYC is sufficient for muscle growth
- PMID: 39482487
- PMCID: PMC11624283
- DOI: 10.1038/s44319-024-00299-z
The 24-hour molecular landscape after exercise in humans reveals MYC is sufficient for muscle growth
Abstract
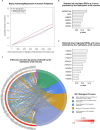
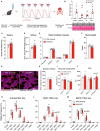
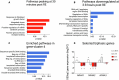
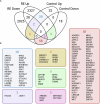
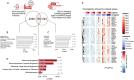
A detailed understanding of molecular responses to a hypertrophic stimulus in skeletal muscle leads to therapeutic advances aimed at promoting muscle mass. To decode the molecular factors regulating skeletal muscle mass, we utilized a 24-h time course of human muscle biopsies after a bout of resistance exercise. Our findings indicate: (1) the DNA methylome response at 30 min corresponds to upregulated genes at 3 h, (2) a burst of translation- and transcription-initiation factor-coding transcripts occurs between 3 and 8 h, (3) changes to global protein-coding gene expression peaks at 8 h, (4) ribosome-related genes dominate the mRNA landscape between 8 and 24 h, (5) methylation-regulated MYC is a highly influential transcription factor throughout recovery. To test whether MYC is sufficient for hypertrophy, we periodically pulse MYC in skeletal muscle over 4 weeks. Transient MYC increases muscle mass and fiber size in the soleus of adult mice. We present a temporally resolved resource for understanding molecular adaptations to resistance exercise in muscle ( http://data.myoanalytics.com ) and suggest that controlled MYC doses influence the exercise-related hypertrophic transcriptional landscape.
Keywords: Biopsy; Methylome; Time Course; Transcription Factors; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. YW is the founder of MyoAnalytics LLC. The authors have no other conflicts to declare.
Figures






References
-
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S (2003) Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev Cell 5:379–390 - PubMed
-
- Alway SE (1997) Overload-induced C-Myc oncoprotein is reduced in aged skeletal muscle. J Gerontol Ser A 52:B203–B211 - PubMed
MeSH terms
Substances
Grants and funding
- K99 AR081367/AR/NIAMS NIH HHS/United States
- R00 AR081367/AR/NIAMS NIH HHS/United States
- P2024-0166/Centrum för Idrottsforskning
- 23137/AMF-Telethon
- AG080047/HHS | National Institutes of Health (NIH)
- AG063994/HHS | National Institutes of Health (NIH)
- P2024-0102)./Centrum för Idrottsforskning
- P20GM139768/The Arkansas Integrated Metabolic Research Center
- P20 GM139768/GM/NIGMS NIH HHS/United States
- P2023-0137/Centrum för Idrottsforskning
- AR081367/HHS | National Institutes of Health (NIH)
- R00 AG063994/AG/NIA NIH HHS/United States
- 2022-01392/Vetenskapsrådet
- R01 AG080047/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

