Cell-autonomous IL6ST activation suppresses prostate cancer development via STAT3/ARF/p53-driven senescence and confers an immune-active tumor microenvironment
- PMID: 39482716
- PMCID: PMC11526557
- DOI: 10.1186/s12943-024-02114-8
Cell-autonomous IL6ST activation suppresses prostate cancer development via STAT3/ARF/p53-driven senescence and confers an immune-active tumor microenvironment
Abstract
Background: Prostate cancer ranks as the second most frequently diagnosed cancer in men worldwide. Recent research highlights the crucial roles IL6ST-mediated signaling pathways play in the development and progression of various cancers, particularly through hyperactivated STAT3 signaling. However, the molecular programs mediated by IL6ST/STAT3 in prostate cancer are poorly understood.
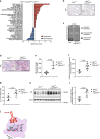
Methods: To investigate the role of IL6ST signaling, we constitutively activated IL6ST signaling in the prostate epithelium of a Pten-deficient prostate cancer mouse model in vivo and examined IL6ST expression in large cohorts of prostate cancer patients. We complemented these data with in-depth transcriptomic and multiplex histopathological analyses.
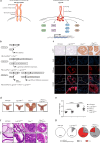
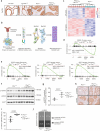
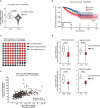
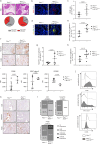
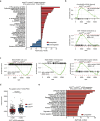
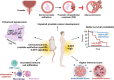
Results: Genetic cell-autonomous activation of the IL6ST receptor in prostate epithelial cells triggers active STAT3 signaling and significantly reduces tumor growth in vivo. Mechanistically, genetic activation of IL6ST signaling mediates senescence via the STAT3/ARF/p53 axis and recruitment of cytotoxic T-cells, ultimately impeding tumor progression. In prostate cancer patients, high IL6ST mRNA expression levels correlate with better recurrence-free survival, increased senescence signals and a transition from an immune-cold to an immune-hot tumor.
Conclusions: Our findings demonstrate a context-dependent role of IL6ST/STAT3 in carcinogenesis and a tumor-suppressive function in prostate cancer development by inducing senescence and immune cell attraction. We challenge the prevailing concept of blocking IL6ST/STAT3 signaling as a functional prostate cancer treatment and instead propose cell-autonomous IL6ST activation as a novel therapeutic strategy.
Keywords: Cytotoxic T-cells; IL6ST/STAT3 signaling; Immune cell infiltration; L-gp130; Prostate cancer; Senescence; Senescence-associated secretory phenotype; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- del Pino-Sedeño T, Infante-Ventura D, de Armas CA, de Pablos-Rodríguez P, Rueda-Domínguez A, Serrano-Aguilar P, Trujillo-Martín MM. Molecular Biomarkers for the Detection of Clinically Significant Prostate Cancer: A Systematic Review and Meta-analysis. Eur Urol Open Sci. 2022;46:105–27. - PMC - PubMed
-
- Mora LB, Buettner R, Seigne J, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: Direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–66. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

