Proteomic analysis of APOEε4 carriers implicates lipid metabolism, complement and lymphocyte signaling in cognitive resilience
- PMID: 39482741
- PMCID: PMC11526661
- DOI: 10.1186/s13024-024-00772-2
Proteomic analysis of APOEε4 carriers implicates lipid metabolism, complement and lymphocyte signaling in cognitive resilience
Abstract
Background: Apolipoprotein E (APOE) ε4 allele is the strongest genetic risk factor for late onset Alzheimer's disease (AD). This case-cohort study used targeted plasma biomarkers and large-scale proteomics to examine the biological mechanisms that allow some APOEε4 carriers to maintain normal cognitive functioning in older adulthood.
Methods: APOEε4 carriers and APOEε3 homozygotes enrolled in the Women's Health Initiative Memory Study (WHIMS) from 1996 to 1999 were classified as resilient if they remained cognitively unimpaired beyond age 80, and as non-resilient if they developed cognitive impairment before or at age 80. AD pathology (Aß42/40) and neurodegeneration (NfL, tau) biomarkers, as well as 1007 proteins (Olink) were quantified in blood collected at study enrollment (on average 14 years prior) when participants were cognitively normal. We identified plasma proteins that distinguished between resilient and non-resilient APOEε4 carriers, examined whether these associations generalized to APOEε3 homozygotes, and replicated these findings in the UK Biobank.
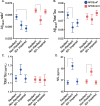
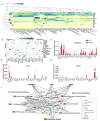
Results: A total of 1610 participants were included (baseline age: 71.3 [3.8 SD] years; all White; 42% APOEε4 carriers). Compared to resilient APOEε4 carriers, non-resilient APOEε4 carriers had lower Aß42/40/tau ratio and greater NfL at baseline. Proteomic analyses identified four proteins differentially expressed between resilient and non-resilient APOEε4 carriers at an FDR-corrected P < 0.05. While one of the candidate proteins, a marker of neuronal injury (NfL), also distinguished resilient from non-resilient APOEε3 homozygotes, the other three proteins, known to be involved in lipid metabolism (ANGPTL4) and immune signaling (PTX3, NCR1), only predicted resilient vs. non-resilient status among APOEε4 carriers (protein*genotype interaction-P < 0.05). Three of these four proteins also predicted 14-year dementia risk among APOEε4 carriers in the UK Biobank validation sample (N = 9420). While the candidate proteins showed little to no association with targeted biomarkers of AD pathology, protein network and enrichment analyses suggested that natural killer (NK) cell and T lymphocyte signaling (via PKC-θ) distinguished resilient from non-resilient APOEε4 carriers.
Conclusions: We identified and replicated a plasma proteomic signature associated with cognitive resilience among APOEε4 carriers. These proteins implicate specific immune processes in the preservation of cognitive status despite elevated genetic risk for AD. Future studies in diverse cohorts will be needed to assess the generalizability of these results.
Keywords: APOE, Resilience; Alzheimer’s disease; Cognition; Immunity; Lipids; Proteomics.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare they have no competing interests.
Figures




References
-
- 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. Alzheimers Dement; 2023;19:1598–695. - PubMed
-
- Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Sci (80-). 1993;261:921–3. - PubMed
-
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet Nat Genet. 2007;39:17–23. - PubMed
-
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet Nat Genet. 1994;7:180–4. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

