Insights into avian molecular cytogenetics-with reptilian comparisons
- PMID: 39482771
- PMCID: PMC11526677
- DOI: 10.1186/s13039-024-00696-y
Insights into avian molecular cytogenetics-with reptilian comparisons
Erratum in
-
Correction: Insights into avian molecular cytogenetics-with reptilian comparisons.Mol Cytogenet. 2024 Nov 22;17(1):29. doi: 10.1186/s13039-024-00699-9. Mol Cytogenet. 2024. PMID: 39578892 Free PMC article. No abstract available.
Abstract
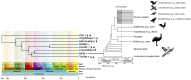
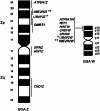
In last 100 years or so, much information has been accumulated on avian karyology, genetics, physiology, biochemistry and evolution. The chicken genome project generated genomic resources used in comparative studies, elucidating fundamental evolutionary processes, much of it funded by the economic importance of domestic fowl (which are also excellent model species in many areas). Studying karyotypes and whole genome sequences revealed population processes, evolutionary biology, and genome function, uncovering the role of repetitive sequences, transposable elements and gene family expansion. Knowledge of the function of many genes and non-expressed or identified regulatory components is however still lacking. Birds (Aves) are diverse, have striking adaptations for flight, migration and survival and inhabit all continents most islands. They also have a unique karyotype with ~ 10 macrochromosomes and ~ 30 microchromosomes that are smaller than other reptiles. Classified into Palaeognathae and Neognathae they are evolutionarily close, and a subset of reptiles. Here we overview avian molecular cytogenetics with reptilian comparisons, shedding light on their karyotypes and genome structure features. We consider avian evolution, then avian (followed by reptilian) karyotypes and genomic features. We consider synteny disruptions, centromere repositioning, and repetitive elements before turning to comparative avian and reptilian genomics. In this context, we review comparative cytogenetics and genome mapping in birds as well as Z- and W-chromosomes and sex determination. Finally, we give examples of pivotal research areas in avian and reptilian cytogenomics, particularly physical mapping and map integration of sex chromosomal genes, comparative genomics of chicken, turkey and zebra finch, California condor cytogenomics as well as some peculiar cytogenetic and evolutionary examples. We conclude that comparative molecular studies and improving resources continually contribute to new approaches in population biology, developmental biology, physiology, disease ecology, systematics, evolution and phylogenetic systematics orientation. This also produces genetic mapping information for chromosomes active in rearrangements during the course of evolution. Further insights into mutation, selection and adaptation of vertebrate genomes will benefit from these studies including physical and online resources for the further elaboration of comparative genomics approaches for many fundamental biological questions.
Keywords: Avian; Bird; Comparative genomics; Cytogenetics; Cytogenomics; Evolution; Genome; Reptile; Sex chromosomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. 10.1038/nature03154. - PubMed
-
- Wallis JW, Aerts J, Groenen M, Crooijmans R, Layman D, Graves T, et al. A physical map of the chicken genome. Nature. 2004;432:761–4. 10.1038/nature03030. - PubMed
-
- Griffin DK, Romanov MN, O’Connor R, Fowler KE, Larkin DM. Avian cytogenetics goes functional. In: Schmid M, Smith J, Burt DW, editors. Third Report on Chicken Genes and Chromosomes 2015. Cytogenet Genome Res. 2015;145:100–5. 10.1159/000430927
Publication types
Grants and funding
- BB/K008226/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 24/2551-0001269-9/Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul
- N/A/International SciKU Branding (ISB), Faculty of Science and Kasetsart University
- N/A/International SciKU Branding (ISB), Faculty of Science and Kasetsart University
LinkOut - more resources
Full Text Sources
Medical

