Sex dimorphism and tissue specificity of gene expression changes in aging mice
- PMID: 39482778
- PMCID: PMC11529319
- DOI: 10.1186/s13293-024-00666-4
Sex dimorphism and tissue specificity of gene expression changes in aging mice
Abstract
Background: Aging is a complex process that involves all tissues in an organism and shows sex dimorphism. While transcriptional changes in aging have been well characterized, the majority of studies have focused on a single sex and sex differences in gene expression in aging are poorly understood. In this study, we explore sex dimorphism in gene expression in aging mice across three tissues.
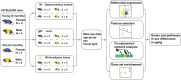
Methods: We collected gastrocnemius muscle, liver and white adipose tissue from young (6 months, n = 14) and old (24 months, n = 14) female and male C57BL/6NIA mice and performed RNA-seq. To investigate sex dimorphism in aging, we considered two levels of comparisons: (a) differentially expressed genes between females and males in the old age group and (b) comparisons between females and males across the aging process. We utilized differential expression analysis and gene feature selection to investigate candidate genes. Gene set enrichment analysis was performed to identify candidate molecular pathways. Furthermore, we performed a co-expression network analysis and chose the gene module(s) associated with aging independent of sex or tissue-type.
Results: We identified both tissue-specific and tissue-independent genes associated with sex dimorphism in aged mice. Unique differentially expressed genes between old males and females across tissues were mainly enriched for pathways related to specific tissue function. We found similar results when exploring sex differences in the aging process, with the exception that in the liver genes enriched for lipid metabolism and digestive system were identified in both females and males. Combining enriched pathways across analyses, we identified amino acid metabolism, digestive system, and lipid metabolism as the core mechanisms of sex dimorphism in aging. Although the vast majority of age-related genes were sex and tissue specific, we identified 127 hub genes contributing to aging independent of sex and tissue that were enriched for the immune system and signal transduction.
Conclusions: There are clear sex differences in gene expression in aging across liver, muscle and white adipose. Core pathways, including amino acid metabolism, digestive system and lipid metabolism, contribute to sex differences in aging.
Keywords: Adipose tissue; Aging; Co-expression network analysis; Feature selection; Gene expression; Liver; Mice; Muscle; Sex dimorphism; Tissue-specific.
Plain language summary
Aging is a complex process that occurs differently across tissues, and in men compared to women. However, the mechanisms that cause sex differences are not well understood. Using naturally aging mouse models we compared how specific genes were differently expressed in muscle, liver and fat of old and young female and male mice. We found that the vast majority of genes that were changed with age were only changed in one sex and specific tissues. Overall, sex differences in aging across tissues were related to genes involved in amino acid metabolism, digestive system and lipid metabolism. Notably, lipid metabolism is important in aging females across all tissues. We also identified a set of genes associated with aging independent of sex and tissue-type involved in immune pathways and signaling. These results enhance our understanding of sex differences in aging.
© 2024. The Author(s).
Conflict of interest statement
D.A.S. is a founder, equity owner, advisor to, director of, board member of, consultant to, investor in and/or inventor on patents licensed to Revere Biosensors, UpRNA, GlaxoSmithKline, Wellomics, DaVinci Logic, InsideTracker (Segterra), Caudalie, Animal Biosciences, Longwood Fund, Catalio Capital Management, Frontier Acquisition Corporation, AFAR (American Federation for Aging Research), Life Extension Advocacy Foundation (LEAF), Cohbar, Galilei, EMD Millipore, Zymo Research, Immetas, Bayer Crop Science, EdenRoc Sciences (and affiliates Arc-Bio, Dovetail Genomics, Claret Bioscience, MetroBiotech, Astrea, Liberty Biosecurity and Delavie), Life Biosciences, Alterity, ATAI Life Sciences, Levels Health, Tally (aka Longevity Sciences) and Bold Capital. D.A.S. is an inventor on a patent application filed by Mayo Clinic and Harvard Medical School that has been licensed to Elysium Health. Additional info on D.A.S. affiliations can be found at
Figures




References
-
- Troen BR. The biology of aging. Mt Sinai J Med. 2003;70:3–22. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

