Circular RNA landscape in extracellular vesicles from human biofluids
- PMID: 39482783
- PMCID: PMC11526565
- DOI: 10.1186/s13073-024-01400-w
Circular RNA landscape in extracellular vesicles from human biofluids
Abstract
Background: Circular RNAs (circRNAs) have emerged as a prominent class of covalently closed single-stranded RNA molecules that exhibit tissue-specific expression and potential as biomarkers in extracellular vesicles (EVs) derived from liquid biopsies. Still, their characteristics and applications in EVs remain to be unveiled.
Methods: We performed a comprehensive analysis of EV-derived circRNAs (EV-circRNAs) using transcriptomics data obtained from 1082 human body fluids, including plasma, urine, cerebrospinal fluid (CSF), and bile. Our validation strategy utilized RT-qPCR and RNA immunoprecipitation assays, complemented by computational techniques for analyzing EV-circRNA features and RNA-binding protein interactions.
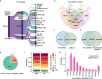
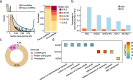
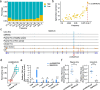
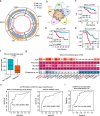
Results: We identified 136,327 EV-circRNAs from various human body fluids. Significantly, a considerable amount of circRNAs with a high back-splicing ratio are highly enriched in EVs compared to linear RNAs. Additionally, we discovered brain-specific circRNAs enriched in plasma EVs and cancer-associated EV-circRNAs linked to clinical outcomes. Moreover, we demonstrated that EV-circRNAs have the potential to serve as biomarkers for evaluating immunotherapy efficacy in non-small cell lung cancer (NSCLC). Importantly, we identified the involvement of RBPs, particularly YBX1, in the sorting mechanism of circRNAs into EVs.
Conclusions: This study unveils the extensive repertoire of EV-circRNAs across human biofluids, offering insights into their potential as disease biomarkers and their mechanistic roles within EVs. The identification of specific circRNAs and the elucidation of RBP-mediated sorting mechanisms open new avenues for the clinical application of EV-circRNAs in disease diagnostics and therapeutics.
Keywords: Biomarker; Cancer; Circular RNAs; Extracellular vesicles; RNA-binding proteins.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Exploring Circular RNA Profile and Expression in Extracellular Vesicles.Methods Mol Biol. 2024;2765:47-59. doi: 10.1007/978-1-0716-3678-7_3. Methods Mol Biol. 2024. PMID: 38381333
-
Altered circular RNA expressions in extracellular vesicles from bronchoalveolar lavage fluids in mice after bacterial infections.Front Immunol. 2024 Apr 4;15:1354676. doi: 10.3389/fimmu.2024.1354676. eCollection 2024. Front Immunol. 2024. PMID: 38638425 Free PMC article.
-
exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids.Nucleic Acids Res. 2022 Jan 7;50(D1):D118-D128. doi: 10.1093/nar/gkab1085. Nucleic Acids Res. 2022. PMID: 34918744 Free PMC article.
-
RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins?J Extracell Vesicles. 2020 Dec;10(2):e12043. doi: 10.1002/jev2.12043. Epub 2020 Dec 28. J Extracell Vesicles. 2020. PMID: 33391635 Free PMC article. Review.
-
Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies.Mol Cancer. 2021 Jan 11;20(1):13. doi: 10.1186/s12943-020-01298-z. Mol Cancer. 2021. PMID: 33430880 Free PMC article. Review.
Cited by
-
Defining the Parameters for Sorting of RNA Cargo Into Extracellular Vesicles.J Extracell Vesicles. 2025 Jul;14(7):e70113. doi: 10.1002/jev2.70113. J Extracell Vesicles. 2025. PMID: 40620070 Free PMC article.
-
Epigenetic Insights into Substance Use Disorder and Associated Psychiatric Conditions.Complex Psychiatry. 2025 Mar 3;11(1):12-36. doi: 10.1159/000544912. eCollection 2025 Jan-Dec. Complex Psychiatry. 2025. PMID: 40201238 Review.
-
Circular RNA discovery with emerging sequencing and deep learning technologies.Nat Genet. 2025 May;57(5):1089-1102. doi: 10.1038/s41588-025-02157-7. Epub 2025 Apr 17. Nat Genet. 2025. PMID: 40247051 Review.
-
Development and validation of a predictive model based upon extracellular vesicle-derived transposable elements for non-invasive detection of pancreatic adenocarcinoma.Biomark Res. 2025 Apr 5;13(1):54. doi: 10.1186/s40364-025-00770-6. Biomark Res. 2025. PMID: 40188348 Free PMC article.
-
Circular RNAs as disease modifiers of complex neurologic disorders.Front Pharmacol. 2025 May 16;16:1577496. doi: 10.3389/fphar.2025.1577496. eCollection 2025. Front Pharmacol. 2025. PMID: 40453669 Free PMC article. Review.
References
-
- Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185(12):2016–34. 10.1016/j.cell.2022.04.021. - PubMed
-
- Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. 10.1038/s41571-021-00585-y. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

