CHAMP delivers accurate taxonomic profiles of the prokaryotes, eukaryotes, and bacteriophages in the human microbiome
- PMID: 39483755
- PMCID: PMC11524946
- DOI: 10.3389/fmicb.2024.1425489
CHAMP delivers accurate taxonomic profiles of the prokaryotes, eukaryotes, and bacteriophages in the human microbiome
Abstract
Introduction: Accurate taxonomic profiling of the human microbiome composition is crucial for linking microbial species to health outcomes. Therefore, we created the Clinical Microbiomics Human Microbiome Profiler (CHAMP), a comprehensive tool designed for the profiling of prokaryotes, eukaryotes, and viruses across all body sites.
Methods: CHAMP uses a reference database derived from 30,382 human microbiome samples, covering 6,567 prokaryotic and 244 eukaryotic species, as well as 64,003 viruses. We benchmarked CHAMP against established profiling tools (MetaPhlAn 4, Bracken 2, mOTUs 3, and Phanta) using a diverse set of in silico metagenomes and DNA mock communities.
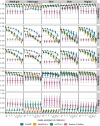
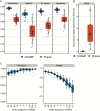
Results: CHAMP demonstrated unparalleled species recall, F1 score, and significantly reduced false positives compared to all other tools benchmarked. The false positive relative abundance (FPRA) for CHAMP was, on average, 50-fold lower than the second-best performing profiler. CHAMP also proved to be more robust than other tools at low sequencing depths, highlighting its application for low biomass samples.
Discussion: Taken together, this establishes CHAMP as a best-in-class human microbiome profiler of prokaryotes, eukaryotes, and viruses in diverse and complex communities across low and high biomass samples. CHAMP profiling is offered as a service by Clinical Microbiomics A/S and is available for a fee at https://cosmosidhub.com.
Keywords: MAG; bacteriophages; benchmarking; human microbiome; metagenomics; taxonomic profiling.
Copyright © 2024 Pita, Myers, Johansen, Russel, Nielsen, Eklund and Nielsen.
Conflict of interest statement
All authors are current or previous employees and/or shareholders of Clinical Microbiomics A/S, a contract research organization offering CHAMP profiling as a service.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

