COADREADx: A comprehensive algorithmic dissection of colorectal cancer unravels salient biomarkers and actionable insights into its discrete progression
- PMID: 39484215
- PMCID: PMC11526798
- DOI: 10.7717/peerj.18347
COADREADx: A comprehensive algorithmic dissection of colorectal cancer unravels salient biomarkers and actionable insights into its discrete progression
Abstract
Background: Colorectal cancer is a common condition with an uncommon burden of disease, heterogeneity in manifestation, and no definitive treatment in the advanced stages. Renewed efforts to unravel the genetic drivers of colorectal cancer progression are paramount. Early-stage detection contributes to the success of cancer therapy and increases the likelihood of a favorable prognosis. Here, we have executed a comprehensive computational workflow aimed at uncovering the discrete stagewise genomic drivers of colorectal cancer progression.
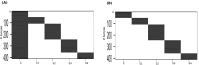
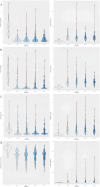
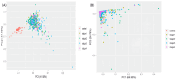
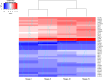
Methods: Using the TCGA COADREAD expression data and clinical metadata, we constructed stage-specific linear models as well as contrast models to identify stage-salient differentially expressed genes. Stage-salient differentially expressed genes with a significant monotone trend of expression across the stages were identified as progression-significant biomarkers. The stage-salient genes were benchmarked using normals-augmented dataset, and cross-referenced with existing knowledge. The candidate biomarkers were used to construct the feature space for learning an optimal model for the digital screening of early-stage colorectal cancers. The candidate biomarkers were also examined for constructing a prognostic model based on survival analysis.
Results: Among the biomarkers identified are: CRLF1, CALB2, STAC2, UCHL1, KCNG1 (stage-I salient), KLHL34, LPHN3, GREM2, ADCY5, PLAC2, DMRT3 (stage-II salient), PIGR, HABP2, SLC26A9 (stage-III salient), GABRD, DKK1, DLX3, CST6, HOTAIR (stage-IV salient), and CDH3, KRT80, AADACL2, OTOP2, FAM135B, HSP90AB1 (top linear model genes). In particular the study yielded 31 genes that are progression-significant such as ESM1, DKK1, SPDYC, IGFBP1, BIRC7, NKD1, CXCL13, VGLL1, PLAC1, SPERT, UPK2, and interestingly three members of the LY6G6 family. Significant monotonic linear model genes included HIGD1A, ACADS, PEX26, and SPIB. A feature space of just seven biomarkers, namely ESM1, DHRS7C, OTOP3, AADACL2, LPHN3, GABRD, and LPAR1, was sufficient to optimize a RandomForest model that achieved > 98% balanced accuracy (and performant recall) of cancer vs. normal on external validation. Design of an optimal multivariate model based on survival analysis yielded a prognostic panel of three stage-IV salient genes, namely HOTAIR, GABRD, and DKK1. Based on the above sparse signatures, we have developed COADREADx, a web-server for potentially assisting colorectal cancer screening and patient risk stratification. COADREADx provides uncertainty measures for its predictions and needs clinical validation. It has been deployed for experimental non-commercial use at: https://apalanialab.shinyapps.io/coadreadx/.
Keywords: Colorectal cancer screening; Differentially expressed genes; Monotonically expressed genes; Network analysis; Progression-significant genes; Random forest; Risk stratification; Stage-salient genes; Stagewise linear models; Web-server.
©2024 Palaniappan et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures





References
-
- Abudoureyimu A, Maimaiti R, Magaoweiya S, Bagedati D, Wen H. Identification of long non-coding RNA expression profile in tissue and serum of papillary thyroid carcinoma. International Journal of Clinical and Experimental Pathology. 2016;9(2):1177–1185.
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA: A Cancer Journal for Clinicians. 2017;67(2):93–99. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, LeKaye HC, Koutcher JA, Cardiff RD, Califano A, Shen MM, Abate-Shen C. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(37):E3506–E3515. - PMC - PubMed
-
- Barret T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

