This is a preprint.
Deletions Rate-Limit Breast and Ovarian Cancer Initiation
- PMID: 39484366
- PMCID: PMC11526986
- DOI: 10.1101/2024.10.17.618945
Deletions Rate-Limit Breast and Ovarian Cancer Initiation
Abstract
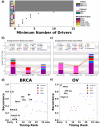
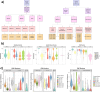
Optimizing prevention and early detection of cancer requires understanding the number, types and timing of driver mutations. To quantify this, we exploited the elevated cancer incidence and mutation rates in germline BRCA1 and BRCA2 (gBRCA1/2) carriers. Using novel statistical models, we identify genomic deletions as the likely rate-limiting mutational processes, with 1-3 deletions required to initiate breast and ovarian tumors. gBRCA1/2-driven hereditary and sporadic tumors undergo convergent evolution to develop a similar set of driver deletions, and deletions explain the elevated cancer risk of gBRCA1/2-carriers. Orthogonal mutation timing analysis identifies deletions of chromosome 17 and 13q as early, recurrent events. Single-cell analyses confirmed deletion rate differences in gBRCA1/2 vs. non-carrier tumors as well as cells engineered to harbor gBRCA1/2. The centrality of deletion-associated chromosomal instability to tumorigenesis shapes interpretation of the somatic evolution of non-malignant tissue and guides strategies for precision prevention and early detection.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS K.H. is a co-founder and board member of a not-for-profit organization, Open Box Science, where he does not receive any compensation. P.C.B. sits on the scientific advisory boards of BioSymetrics, Inc. and Intersect Diagnostics, Inc. and previously sat on that of Sage Bionetworks. All other authors declare no competing financial interests.
Figures



Similar articles
-
Endometrial Cancers in BRCA1 or BRCA2 Germline Mutation Carriers: Assessment of Homologous Recombination DNA Repair Defects.JCO Precis Oncol. 2019 Aug 29;3:PO.19.00103. doi: 10.1200/PO.19.00103. eCollection 2019. JCO Precis Oncol. 2019. PMID: 32914019 Free PMC article.
-
Mutations of TP53 and genes related to homologous recombination repair in breast cancer with germline BRCA1/2 mutations.Hum Genomics. 2023 Jan 6;17(1):2. doi: 10.1186/s40246-022-00447-3. Hum Genomics. 2023. PMID: 36604691 Free PMC article.
-
Somatic BRCA1/2 mutations are associated with a similar survival advantage to their germline counterparts in tubo-ovarian high grade serous carcinoma.Eur J Cancer. 2025 Mar 26;219:115299. doi: 10.1016/j.ejca.2025.115299. Epub 2025 Feb 11. Eur J Cancer. 2025. PMID: 39955805
-
Hematologic toxicities of chemotherapy in breast and ovarian cancer patients carrying BRCA1/BRCA2 germline pathogenic variants. A single center experience and review of the literature.Fam Cancer. 2023 Jul;22(3):283-289. doi: 10.1007/s10689-023-00331-6. Epub 2023 Apr 29. Fam Cancer. 2023. PMID: 37119509 Free PMC article. Review.
-
BRCA1 and BRCA2 and the genetics of breast and ovarian cancer.Hum Mol Genet. 2001 Apr;10(7):705-13. doi: 10.1093/hmg/10.7.705. Hum Mol Genet. 2001. PMID: 11257103 Review.
References
-
- Garraway L. A., Lander E. S., Lessons from the cancer genome. Cell 153, 17–37 (2013). - PubMed
-
- Little M. P., Are two mutations sufficient to cause cancer? Some generalizations of the two-mutation model of carcinogenesis of Moolgavkar, Venzon, and Knudson, and of the multistage model of Armitage and Doll. Biometrics 51, 1278–1291 (1995). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
