This is a preprint.
The NHGRI-EBI GWAS Catalog: standards for reusability, sustainability and diversity
- PMID: 39484403
- PMCID: PMC11526975
- DOI: 10.1101/2024.10.23.619767
The NHGRI-EBI GWAS Catalog: standards for reusability, sustainability and diversity
Update in
-
The NHGRI-EBI GWAS Catalog: standards for reusability, sustainability and diversity.Nucleic Acids Res. 2025 Jan 6;53(D1):D998-D1005. doi: 10.1093/nar/gkae1070. Nucleic Acids Res. 2025. PMID: 39530240 Free PMC article.
Abstract
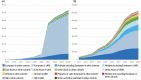
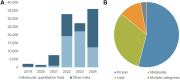
The NHGRI-EBI GWAS Catalog serves as a vital resource for the genetic research community, providing access to the most comprehensive database of human GWAS results. Currently, it contains close to 7,000 publications for more than 15,000 traits, from which more than 625,000 lead associations have been curated. Additionally, 85,000 full genome-wide summary statistics datasets - containing association data for all variants in the analysis - are available for downstream analyses such as meta-analysis, fine-mapping, Mendelian randomisation or development of polygenic risk scores. As a centralised repository for GWAS results, the GWAS Catalog sets and implements standards for data submission and harmonisation, and encourages the use of consistent descriptors for traits, samples and methodologies. We share processes and vocabulary with the PGS Catalog, improving interoperability for a growing user group. Here, we describe the latest changes in data content, improvements in our user interface, and the implementation of the GWAS-SSF standard format for summary statistics. We address the challenges of handling the rapid increase in large-scale molecular quantitative trait GWAS and the need for sensitivity in the use of population and cohort descriptors while maintaining data interoperability and reusability.
Conflict of interest statement
Conflict of Interest statement: M.I. is a trustee of the Public Health Genomics (PHG) Foundation, a member of the Scientific Advisory Board of Open Targets, and has research collaborations with AstraZeneca, Nightingale Health and Pfizer which are unrelated to this study. No other conflicts of interest were reported by authors.
Figures

References
-
- Mallard T.T., Linner R.K., Grotzinger A.D., Sanchez-Roige S., Seidlitz J., Okbay A., de Vlaming R., Meddens S.F.W., Bipolar Disorder Working Group of the Psychiatric Genomics, C., Palmer A.A. et al. (2022) Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genom, 2. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
