This is a preprint.
Integrin alpha1 beta1 promotes interstitial fibrosis in a mouse model of polycystic kidney disease
- PMID: 39484448
- PMCID: PMC11526950
- DOI: 10.1101/2024.10.18.619080
Integrin alpha1 beta1 promotes interstitial fibrosis in a mouse model of polycystic kidney disease
Abstract
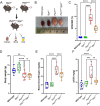
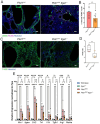
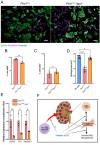
Fibrosis is the cause of end-stage kidney failure in patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD). The molecular and cellular mechanisms involved in fibrosis are complex and anti-fibrotic therapies have so far failed to make an impact on patient welfare. Using unbiased proteomics analysis on the Pkd1 nl/nl mouse, we found that expression of the integrin α1 subunit is increased in this model of ADPKD. In human ADPKD tissue and two single cell RNA kidney disease datasets, ITGA1 was also upregulated. To investigate the functional role of this integrin subunit in ADPKD, we generated a Pkd1 nl/nl Itga1 -/- mouse. We observed a significant reduction in kidney volume and kidney dysfunction in mice lacking the integrin α1 subunit. Kidneys from Pkd1 nl/nl Itga1 -/- mice had smaller cysts and reduced interstitial expansion and tubular atrophy. Picrosirius red staining identified a restriction in collagen staining in the interstitium and the myofibroblast marker α smooth muscle actin was also downregulated. Myofibroblast cell proliferation was reduced in Pkd1 nl/nl Itga1 -/- mice and primary fibroblast cultures demonstrated an abrogated fibrogenic phenotype in integrin α1-depleted fibroblasts. These results highlight a previously unrecognised role for the integrin α1 subunit in kidney fibrosis.
Figures





References
-
- Hughes J., et al., The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet, 1995. 10(2): p. 151–60. - PubMed
-
- Grieben M., et al., Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2). Nat Struct Mol Biol, 2017. 24(2): p. 114–122. - PubMed
-
- Su Q., et al., Structure of the human PKD1-PKD2 complex. Science, 2018. 361(6406). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
