This is a preprint.
Establishing the green algae Chlamydomonas incerta as a platform for recombinant protein production
- PMID: 39484490
- PMCID: PMC11527144
- DOI: 10.1101/2024.10.25.618925
Establishing the green algae Chlamydomonas incerta as a platform for recombinant protein production
Update in
-
Engineering the green algae Chlamydomonas incerta for recombinant protein production.PLoS One. 2025 Apr 16;20(4):e0321071. doi: 10.1371/journal.pone.0321071. eCollection 2025. PLoS One. 2025. PMID: 40238798 Free PMC article.
Abstract
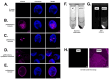
Chlamydomonas incerta, a genetically close relative of the model green alga Chlamydomonas reinhardtii, shows significant potential as a host for recombinant protein expression. Because of the close genetic relationship between C. incerta and C. reinhardtii, this species offers an additional reference point for advancing our understanding of photosynthetic organisms, and also provides a potential new candidate for biotechnological applications. This study investigates C. incerta's capacity to express three recombinant proteins: the fluorescent protein mCherry, the hemicellulose-degrading enzyme xylanase, and the plastic-degrading enzyme PHL7. We have also examined the capacity to target protein expression to various cellular compartments in this alga, including the cytosol, secretory pathway, cytoplasmic membrane, and cell wall. When compared directly with C. reinhardtii, C. incerta exhibited a distinct but notable capacity for recombinant protein production. Cellular transformation with a vector encoding mCherry revealed that C. incerta produced approximately 3.5 times higher fluorescence levels and a 3.7-fold increase in immunoblot intensity compared to C. reinhardtii. For xylanase expression and secretion, both C. incerta and C. reinhardtii showed similar secretion capacities and enzymatic activities, with comparable xylan degradation rates, highlighting the industrial applicability of xylanase expression in microalgae. Finally, C. incerta showed comparable PHL7 activity levels to C. reinhardtii, as demonstrated by the in vitro degradation of a polyester polyurethane suspension, Impranil® DLN. Finally, we also explored the potential of cellular fusion for the generation of genetic hybrids between C. incerta and C. reinhardtii as a means to enhance phenotypic diversity and augment genetic variation. We were able to generate genetic fusion that could exchange both the recombinant protein genes, as well as associated selectable marker genes into recombinant offspring. These findings emphasize C. incerta's potential as a robust platform for recombinant protein production, and as a powerful tool for gaining a better understanding of microalgal biology.
Keywords: Microalgae; biotechnology; cellular localization; hybridization; industrial enzymes; secretion.
Conflict of interest statement
CONFLICT OF INTEREST SM was a founding of and holds an equity stake in Algenesis Inc, a company that could potentially benefit from this work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol [Internet]. 2014. Apr 17 [cited 2024 Oct 12];5. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2014.00172/abstract - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
