This is a preprint.
Foxi1 regulates multiple steps of mucociliary development and ionocyte specification through transcriptional and epigenetic mechanisms
- PMID: 39484493
- PMCID: PMC11527170
- DOI: 10.1101/2024.10.27.620464
Foxi1 regulates multiple steps of mucociliary development and ionocyte specification through transcriptional and epigenetic mechanisms
Abstract
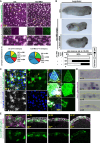
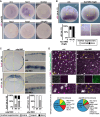
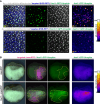
Foxi1 is a master regulator of ionocytes (ISCs / INCs) across species and organs. Two subtypes of ISCs exist, and both α- and β-ISCs regulate pH- and ion-homeostasis in epithelia. Gain and loss of FOXI1 function are associated with human diseases, including Pendred syndrome, male infertility, renal acidosis and cancers. Foxi1 functions were predominantly studied in the context of ISC specification, however, reports indicate additional functions in early and ectodermal development. Here, we re-investigated the functions of Foxi1 in Xenopus laevis embryonic mucociliary epidermis development and found a novel function for Foxi1 in the generation of Notch-ligand expressing mucociliary multipotent progenitors (MPPs). We demonstrate that Foxi1 has multiple concentration-dependent functions: At low levels, Foxi1 confers ectodermal competence through transcriptional and epigenetic mechanisms, while at high levels, Foxi1 induces a multi-step process of ISC specification and differentiation. We further describe how foxi1 expression is affected through auto- and Notch-regulation, how Ubp1 and Dmrt2 regulate ISC subtype differentiation, and how this developmental program affects Notch signaling as well as mucociliary patterning. Together, we reveal novel functions for Foxi1 in Xenopus mucociliary epidermis formation, relevant to our understanding of vertebrate development and human disease.
Keywords: Xenopus epidermis; development; ionocytes; mucociliary; multipotent progenitors.
Figures


Similar articles
-
Temporal Notch signaling regulates mucociliary cell fates through Hes-mediated competitive de-repression.bioRxiv [Preprint]. 2023 Feb 15:2023.02.15.528675. doi: 10.1101/2023.02.15.528675. bioRxiv. 2023. PMID: 36824900 Free PMC article. Preprint.
-
mab21-l3 regulates cell fate specification of multiciliate cells and ionocytes.Nat Commun. 2015 Jan 19;6:6017. doi: 10.1038/ncomms7017. Nat Commun. 2015. PMID: 25598413
-
BMP signalling controls the construction of vertebrate mucociliary epithelia.Development. 2015 Jul 1;142(13):2352-63. doi: 10.1242/dev.118679. Epub 2015 Jun 19. Development. 2015. PMID: 26092849
-
Xenopus epidermal and endodermal epithelia as models for mucociliary epithelial evolution, disease, and metaplasia.Genesis. 2021 Feb;59(1-2):e23406. doi: 10.1002/dvg.23406. Epub 2021 Jan 5. Genesis. 2021. PMID: 33400364 Review.
-
Signaling Control of Mucociliary Epithelia: Stem Cells, Cell Fates, and the Plasticity of Cell Identity in Development and Disease.Cells Tissues Organs. 2022;211(6):736-753. doi: 10.1159/000514579. Epub 2021 Apr 26. Cells Tissues Organs. 2022. PMID: 33902038 Free PMC article. Review.
References
-
- Afouda B.A., Ciau-Uitz A., and Patient R.. 2005. GATA4, 5 and 6 mediate TGFbeta maintenance of endodermal gene expression in Xenopus embryos. Development. 132:763–774. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
