This is a preprint.
Single-chain nanobody inhibition of Notch and avidity enhancement utilizing the β-pore forming toxin Aerolysin
- PMID: 39484510
- PMCID: PMC11526928
- DOI: 10.1101/2024.10.22.617501
Single-chain nanobody inhibition of Notch and avidity enhancement utilizing the β-pore forming toxin Aerolysin
Update in
-
Single-Chain Nanobody Inhibition of Notch and Avidity Enhancement Utilizing the β-Pore-Forming Toxin Aerolysin.ACS Chem Biol. 2025 Mar 21;20(3):656-669. doi: 10.1021/acschembio.4c00803. Epub 2025 Mar 13. ACS Chem Biol. 2025. PMID: 40079390 Free PMC article.
Abstract
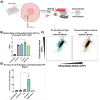
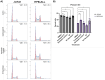
Notch plays critical roles in developmental processes and disease pathogenesis, which has led to numerous efforts to modulate its function with small molecules and antibodies. Here we present a nanobody inhibitor of Notch signaling, derived from a synthetic phage-display library targeting the notch Negative Regulatory Region (NRR). The nanobody inhibits Notch signaling in a luciferase reporter assay and in Notch-dependent hematopoietic progenitor cell differentiation assay, despite a modest 19uM affinity for Notch. We addressed the low affinity by fusion to a membrane-associating domain derived from the β-Pore forming toxin Aerolysin, resulting in a significantly improved IC50 for Notch inhibition. The nanobody-aerolysin fusion inhibits proliferation of T-ALL cell lines with similar efficacy to other Notch pathway inhibitors. Overall, this study reports the development of a Notch inhibitory antibody, and demonstrates a proof-of-concept for a generalizable strategy to increase the efficacy and potency of low-affinity antibody binders.
Conflict of interest statement
Conflict of interest statement The authors declare no conflicts of interest in this work.
Figures




References
-
- Bray S.J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
