The genome sequence of the Brindled Ochre moth, Dasypolia templi (Thunberg, 1792)
- PMID: 39484641
- PMCID: PMC11525101
- DOI: 10.12688/wellcomeopenres.23054.1
The genome sequence of the Brindled Ochre moth, Dasypolia templi (Thunberg, 1792)
Abstract
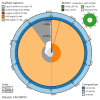
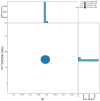
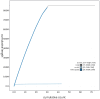
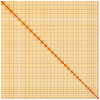
We present a genome assembly from an individual male Brindled Ochre moth, Dasypolia templi (Arthropoda; Insecta; Lepidoptera; Noctuidae). The genome sequence has a total length of 855.30 megabases. Most of the assembly is scaffolded into 31 chromosomal pseudomolecules, including the Z sex chromosome. The mitochondrial genome has also been assembled and is 15.37 kilobases in length.
Keywords: Brindled Ochre moth; Dasypolia templi; Lepidoptera; chromosomal; genome sequence.
Copyright: © 2024 Griffiths A et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
The genome sequence of the Green-brindled Crescent, Allophyes oxyacanthae (Linnaeus, 1758).Wellcome Open Res. 2023 Feb 3;8:53. doi: 10.12688/wellcomeopenres.18935.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39148947 Free PMC article.
-
The genome sequence of the Brindled Flat-body, Agonopterix arenella (Denis & Schiffermüller, 1775).Wellcome Open Res. 2023 May 12;8:214. doi: 10.12688/wellcomeopenres.19252.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39262678 Free PMC article.
-
The genome sequence of the Sandy Carpet moth, Perizoma flavofasciatum (Thunberg, 1792).Wellcome Open Res. 2025 Feb 3;10:40. doi: 10.12688/wellcomeopenres.23612.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40062320 Free PMC article.
-
The genome sequence of the Sprawler moth, Asteroscopus sphinx Hufnagel, 1766.Wellcome Open Res. 2024 Oct 11;9:505. doi: 10.12688/wellcomeopenres.22920.2. eCollection 2024. Wellcome Open Res. 2024. PMID: 39600925 Free PMC article.
-
The genome sequence of the Mouse Moth, Amphipyra tragopoginis (Clerck 1759).Wellcome Open Res. 2023 Feb 3;8:54. doi: 10.12688/wellcomeopenres.18946.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37363059 Free PMC article.
References
-
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 - DOI
-
- Beasley J, Uhl R, Forrest LL, et al. : DNA barcoding SOPs for the Darwin Tree of Life project. protocols.io. 2023; (accessed 25 June 2024). 10.17504/protocols.io.261ged91jv47/v1 - DOI
LinkOut - more resources
Full Text Sources

