Cyanorhodopsin-II represents a yellow-absorbing proton-pumping rhodopsin clade within cyanobacteria
- PMID: 39485071
- PMCID: PMC11528372
- DOI: 10.1093/ismejo/wrae175
Cyanorhodopsin-II represents a yellow-absorbing proton-pumping rhodopsin clade within cyanobacteria
Abstract
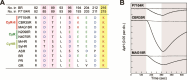
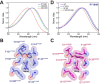
Microbial rhodopsins are prevalent in many cyanobacterial groups as a light-energy-harvesting system in addition to the photosynthetic system. It has been suggested that this dual system allows efficient capture of sunlight energy using complementary ranges of absorption wavelengths. However, the diversity of cyanobacterial rhodopsins, particularly in accumulated metagenomic data, remains underexplored. Here, we used a metagenomic mining approach, which led to the identification of a novel rhodopsin clade unique to cyanobacteria, cyanorhodopsin-II (CyR-II). CyR-IIs function as light-driven outward H+ pumps. CyR-IIs, together with previously identified cyanorhodopsins (CyRs) and cyanobacterial halorhodopsins (CyHRs), constitute cyanobacterial ion-pumping rhodopsins (CyipRs), a phylogenetically distinct family of rhodopsins. The CyR-II clade is further divided into two subclades, YCyR-II and GCyR-II, based on their specific absorption wavelength. YCyR-II absorbed yellow light (λmax = 570 nm), whereas GCyR-II absorbed green light (λmax = 550 nm). X-ray crystallography and mutational analysis revealed that the difference in absorption wavelengths is attributable to slight changes in the side chain structure near the retinal chromophore. The evolutionary trajectory of cyanobacterial rhodopsins suggests that the function and light-absorbing range of these rhodopsins have been adapted to a wide range of habitats with variable light and environmental conditions. Collectively, these findings shed light on the importance of rhodopsins in the evolution and environmental adaptation of cyanobacteria.
Keywords: cyanobacteria; ecology; evolution; microbial rhodopsin.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Miyashita H, Ikemoto H, Kurano Net al. Chlorophyll d as a major pigment. Nature 1996;383:402–2. 10.1038/383402a0 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

