Evidence for a role of human blood-borne factors in mediating age-associated changes in molecular circadian rhythms
- PMID: 39485282
- PMCID: PMC11530234
- DOI: 10.7554/eLife.88322
Evidence for a role of human blood-borne factors in mediating age-associated changes in molecular circadian rhythms
Abstract
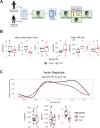
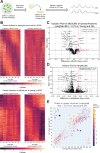
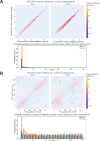
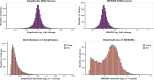
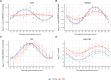
Aging is associated with a number of physiologic changes including perturbed circadian rhythms; however, mechanisms by which rhythms are altered remain unknown. To test the idea that circulating factors mediate age-dependent changes in peripheral rhythms, we compared the ability of human serum from young and old individuals to synchronize circadian rhythms in culture. We collected blood from apparently healthy young (age 25-30) and old (age 70-76) individuals at 14:00 and used the serum to synchronize cultured fibroblasts. We found that young and old sera are equally competent at initiating robust ~24 hr oscillations of a luciferase reporter driven by clock gene promoter. However, cyclic gene expression is affected, such that young and old sera promote cycling of different sets of genes. Genes that lose rhythmicity with old serum entrainment are associated with oxidative phosphorylation and Alzheimer's Disease as identified by STRING and IPA analyses. Conversely, the expression of cycling genes associated with cholesterol biosynthesis increased in the cells entrained with old serum. Genes involved in the cell cycle and transcription/translation remain rhythmic in both conditions. We did not observe a global difference in the distribution of phase between groups, but found that peak expression of several clock-controlled genes (PER3, NR1D1, NR1D2, CRY1, CRY2, and TEF) lagged in the cells synchronized ex vivo with old serum. Taken together, these findings demonstrate that age-dependent blood-borne factors affect circadian rhythms in peripheral cells and have the potential to impact health and disease via maintaining or disrupting rhythms respectively.
Keywords: aging; cell biology; circadian rhythms; human; transcriptomics.
© 2023, Schwarz et al.
Conflict of interest statement
JS, AM, NL, YL, CH, GG, CS, SZ No competing interests declared, AS Reviewing editor, eLife
Figures







Update of
-
Evidence for a role of human blood-borne factors in mediating age-associated changes in molecular circadian rhythms.bioRxiv [Preprint]. 2024 Jul 1:2023.04.19.537477. doi: 10.1101/2023.04.19.537477. bioRxiv. 2024. Update in: Elife. 2024 Nov 01;12:RP88322. doi: 10.7554/eLife.88322. PMID: 37808824 Free PMC article. Updated. Preprint.
References
-
- Bertolotti M, Mussi C, Pellegrini E, Magni A, Del Puppo M, Ognibene S, Carulli L, Anzivino C, Baldelli E, Loria P, Carulli N. Age-associated alterations in cholesterol homeostasis: evidence from a cross-sectional study in a Northern Italy population. Clinical Interventions in Aging. 2014;9:425–432. doi: 10.2147/CIA.S57714. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- UL1 TR001878/TR/NCATS NIH HHS/United States
- T32 NS105607/NS/NINDS NIH HHS/United States
- NIH NS48471/NS/NINDS NIH HHS/United States
- 5UL1TR001878/RR/NCRR NIH HHS/United States
- R01 NS048471/NS/NINDS NIH HHS/United States
- R00 HL147212/HL/NHLBI NIH HHS/United States
- Gilliam Fellowship/HHMI/Howard Hughes Medical Institute/United States
- R37 NS048471/NS/NINDS NIH HHS/United States
- Investigator/HHMI/Howard Hughes Medical Institute/United States
- R00HL147212/HL/NHLBI NIH HHS/United States
- P30 DK019525/DK/NIDDK NIH HHS/United States
- T32-NS105607/NS/NINDS NIH HHS/United States
- R56 NS048471/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

