ATP-competitive inhibitors of PI3K enzymes demonstrate an isoform selective dual action by controlling membrane binding
- PMID: 39485310
- PMCID: PMC7617104
- DOI: 10.1042/BCJ20240479
ATP-competitive inhibitors of PI3K enzymes demonstrate an isoform selective dual action by controlling membrane binding
Abstract
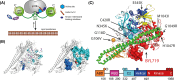
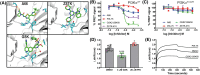
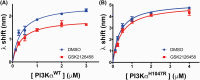
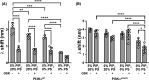
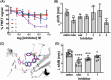
PI3Kα, consisting of the p110α isoform of the catalytic subunit of PI 3-kinase (encoded by PIK3CA) and the p85α regulatory subunit (encoded by PI3KR1) is activated by growth factor receptors. The identification of common oncogenic mutations in PIK3CA has driven the development of many inhibitors that bind to the ATP-binding site in the p110α subunit. Upon activation, PI3Kα undergoes conformational changes that promote its membrane interaction and catalytic activity, yet the effects of ATP-site directed inhibitors on the PI3Kα membrane interaction are unknown. Using FRET and biolayer interferometry assays, we show that a class of ATP-site directed inhibitors represented by GSK2126458 block the growth factor activated PI3KαWT membrane interaction, an activity dependent on the ligand forming specific ATP-site interactions. The membrane interaction for hot spot oncogenic mutations that bypass normal p85α regulatory mechanisms was insensitive to GSK2126458, while GSK2126458 could regulate mutations found outside of these hot spot regions. Our data show that the effect of GSK126458 on the membrane interaction requires the enzyme to revert from its growth factor activated state to a basal state. We find that an ATP substrate analogue can increase the wild type PI3Kα membrane interaction, uncovering a substrate based regulatory event that can be mimicked by different inhibitor chemotypes. Our findings, together with the discovery of small molecule allosteric activators of PI3Kα illustrate that PI3Kα membrane interactions can be modulated by factors related to ligand binding both within the ATP site and at allosteric sites.
Keywords: PIK3CA; lipid kinase; membrane proteins; phosphoinositide 3-kinase; protein conformation; small molecules.
© 2024 The Author(s).
Conflict of interest statement
J.E.B. reports consulting fees from Scorpion Therapeutics, Reactive therapeutics, and Olema Oncology. J.E.B. reports research contracts from Novartis, Olema Oncology, Scorpion Therapeutics and Calico Life Sciences. The other author authors declare that there are no competing interests associated with the manuscript.
Figures
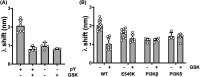
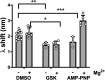
References
-
- Tsolakos, N., Durrant, T.N., Chessa, T., Suire, S.M., Oxley, D., Kulkarni, S.et al. (2018) Quantitation of class IA PI3Ks in mice reveals p110-free-p85s and isoform-selective subunit associations and recruitment to receptors. Proc. Natl Acad. Sci. U.S.A. 115, 12176–12181 10.1073/pnas.1803446115 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

