Identification of the m6A/m5C/m1A methylation modification genes in Alzheimer's disease based on bioinformatic analysis
- PMID: 39485681
- PMCID: PMC11719101
- DOI: 10.18632/aging.206146
Identification of the m6A/m5C/m1A methylation modification genes in Alzheimer's disease based on bioinformatic analysis
Abstract
Background: As a progressive neurodegenerative disease, the comprehensive understanding of the pathogenesis of Alzheimer's disease (AD) is yet to be clarified. Modifications in RNA, including m6A/m5C/m1A, affect the onset and progression of many diseases. Consequently, this study focuses on the role of methylation modification in the pathogenesis of AD.
Materials and methods: Three AD-related datasets, namely GSE33000, GSE122063, and GSE44770, were acquired from GEO. Differential analysis of m6A/m5C/m1A regulator genes was conducted. Applying a consensus clustering approach, distinct subtypes within AD were identified as per the expression patterns of relevant differentially expressed genes. Machine learning models were constructed to identify five significant genes from the best model. The analysis of hub gene-based drug regulatory networks and ceRNA regulatory networks was conducted by Cytoscape.
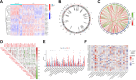
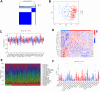
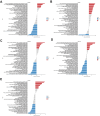
Results: In comparison to non-AD patients, 24 genes were identified as dysregulated in AD patients, and these genes were associated with various immunological characteristics. Two distinct clusters were successfully identified through consensus clustering, with cluster 2 demonstrating higher immune characteristics compared to cluster 1. The performance of four machine learning models was determined by conducting a receiver operating characteristic (ROC) analysis. The analysis revealed that the SVM model achieved the highest AUC value of 0.947. Five genes (YTHDF1, METTL3, DNMT1, DNMT3A, ALKBH1) were selected as the predicted genes. Finally, a hub gene-based Gene-Drug regulatory network and a ceRNA regulatory network were successfully developed.
Conclusions: The findings offered fresh perspectives on the molecular patterns and immune mechanisms underlying AD, contributing valuable insights into our understanding of this complex neurodegenerative disorder.
Keywords: Alzheimer’s disease; cluster analysis; diagnostic model; immune characteristics; machine learning.
Conflict of interest statement
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

