Spatial multiplex analysis of lung cancer reveals that regulatory T cells attenuate KRAS-G12C inhibitor-induced immune responses
- PMID: 39485838
- PMCID: PMC11529713
- DOI: 10.1126/sciadv.adl6464
Spatial multiplex analysis of lung cancer reveals that regulatory T cells attenuate KRAS-G12C inhibitor-induced immune responses
Abstract
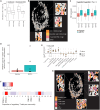
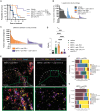
Kirsten rat sarcoma virus (KRAS)-G12C inhibition causes remodeling of the lung tumor immune microenvironment and synergistic responses to anti-PD-1 treatment, but only in T cell infiltrated tumors. To investigate mechanisms that restrain combination immunotherapy sensitivity in immune-excluded tumors, we used imaging mass cytometry to explore cellular distribution in an immune-evasive KRAS mutant lung cancer model. Cellular spatial pattern characterization revealed a community where CD4+ and CD8+ T cells and dendritic cells were gathered, suggesting localized T cell activation. KRAS-G12C inhibition led to increased PD-1 expression, proliferation, and cytotoxicity of CD8+ T cells, and CXCL9 expression by dendritic cells, indicating an effector response. However, suppressive regulatory T cells (Tregs) were also found in frequent contact with effector T cells within this community. Lung adenocarcinoma clinical samples showed similar communities. Depleting Tregs led to enhanced tumor control in combination with anti-PD-1 and KRAS-G12C inhibitor. Combining Treg depletion with KRAS inhibition shows therapeutic potential for increasing antitumoral immune responses.
Figures






References
-
- Borghaei H., Paz-Ares L., Horn L., Spigel D. R., Steins M., Ready N. E., Chow L. Q., Vokes E. E., Felip E., Holgado E., Barlesi F., Kohlhäufl M., Arrieta O., Burgio M. A., Fayette J., Lena H., Poddubskaya E., Gerber D. E., Gettinger S. N., Rudin C. M., Rizvi N., Crinò L., Blumenschein G. R. Jr., Antonia S. J., Dorange C., Harbison C. T., Graf Finckenstein F., Brahmer J. R., Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015). - PMC - PubMed
-
- Gettinger S. N., Wurtz A., Goldberg S. B., Rimm D., Schalper K., Kaech S., Kavathas P., Chiang A., Lilenbaum R., Zelterman D., Politi K., Herbst R. S., Clinical features and management of acquired resistance to PD-1 axis inhibitors in 26 patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 13, 831–839 (2018). - PMC - PubMed
-
- de Langen A. J., Johnson M. L., Mazieres J., Dingemans A. C., Mountzios G., Pless M., Wolf J., Schuler M., Lena H., Skoulidis F., Yoneshima Y., Kim S. W., Linardou H., Novello S., van der Wekken A. J., Chen Y., Peters S., Felip E., Solomon B. J., Ramalingam S. S., Dooms C., Lindsay C. R., Ferreira C. G., Blais N., Obiozor C. C., Wang Y., Mehta B., Varrieur T., Ngarmchamnanrith G., Stollenwerk B., Waterhouse D., Paz-Ares L., CodeBrea K. I., Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: A randomised, open-label, phase 3 trial. Lancet 401, 733–746 (2023). - PubMed
-
- Skoulidis F., Li B. T., Dy G. K., Price T. J., Falchook G. S., Wolf J., Italiano A., Schuler M., Borghaei H., Barlesi F., Kato T., Curioni-Fontecedro A., Sacher A., Spira A., Ramalingam S. S., Takahashi T., Besse B., Anderson A., Ang A., Tran Q., Mather O., Henary H., Ngarmchamnanrith G., Friberg G., Velcheti V., Govindan R., Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 384, 2371–2381 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

