Neural crest precursors from the skin are the primary source of directly reprogrammed neurons
- PMID: 39486406
- PMCID: PMC11589197
- DOI: 10.1016/j.stemcr.2024.10.003
Neural crest precursors from the skin are the primary source of directly reprogrammed neurons
Abstract
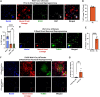
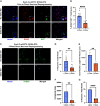
Direct reprogramming involves the conversion of differentiated cell types without returning to an earlier developmental state. Here, we explore how heterogeneity in developmental lineage and maturity of the starting cell population contributes to direct reprogramming using the conversion of murine fibroblasts into neurons. Our hypothesis is that a single lineage of cells contributes to most reprogramming and that a rare elite precursor with intrinsic bias is the source of reprogrammed neurons. We find that nearly all reprogrammed neurons are derived from the neural crest (NC) lineage. Moreover, when rare proliferating NC precursors are selectively ablated, there is a large reduction in the number of reprogrammed neurons. Previous interpretations of this paradigm are that it demonstrates a cell fate conversion across embryonic germ layers (mesoderm to ectoderm). Our interpretation is that this is actually directed differentiation of a neural lineage stem cell in the skin that has intrinsic bias to produce neuronal progeny.
Keywords: cell identity; direct reprogramming; embryonic lineage; fate mapping; germ layer; neural crest; neuron; plasticity; precursor cell differentiation; skin.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

