Genetics of Latin American Diversity Project: Insights into population genetics and association studies in admixed groups in the Americas
- PMID: 39486408
- PMCID: PMC11605695
- DOI: 10.1016/j.xgen.2024.100692
Genetics of Latin American Diversity Project: Insights into population genetics and association studies in admixed groups in the Americas
Abstract
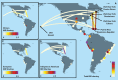
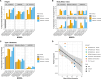
Latin Americans are underrepresented in genetic studies, increasing disparities in personalized genomic medicine. Despite available genetic data from thousands of Latin Americans, accessing and navigating the bureaucratic hurdles for consent or access remains challenging. To address this, we introduce the Genetics of Latin American Diversity (GLAD) Project, compiling genome-wide information from 53,738 Latin Americans across 39 studies representing 46 geographical regions. Through GLAD, we identified heterogeneous ancestry composition and recent gene flow across the Americas. Additionally, we developed GLAD-match, a simulated annealing-based algorithm, to match the genetic background of external samples to our database, sharing summary statistics (i.e., allele and haplotype frequencies) without transferring individual-level genotypes. Finally, we demonstrate the potential of GLAD as a critical resource for evaluating statistical genetic software in the presence of admixture. By providing this resource, we promote genomic research in Latin Americans and contribute to the promises of personalized medicine to more people.
Keywords: GLAD-match; GWAS; Latin America; identity-by-descent; imputation; local ancestry; migration; population structure.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests. D.P.L. is now an employee of AstraZeneca. This is unrelated to the work of this paper.
Figures




References
-
- Manichaikul A., Palmas W., Rodriguez C.J., Peralta C.A., Divers J., Guo X., Chen W.-M., Wong Q., Williams K., Kerr K.F., et al. Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002640. - DOI - PMC - PubMed
-
- Plecher H. Statista; 2019. Latin America - Statistics & Facts.https://www.statista.com/topics/3287/latin-america/
-
- Noe-Bustamante L., Hugo Lopez M., Manuel Krogstad J. Pew Res. Cent. US Hisp. Popul. Surpassed 60 Million 2019 Growth Has Slowed. 2020. U.S. Hispanic population surpassed 60 million in 2019, but growth has slowed.https://www.pewresearch.org/fact-tank/2020/07/07/u-s-hispanic-population...

