Neoadjuvant vidutolimod and nivolumab in high-risk resectable melanoma: A prospective phase II trial
- PMID: 39486411
- PMCID: PMC11560503
- DOI: 10.1016/j.ccell.2024.10.007
Neoadjuvant vidutolimod and nivolumab in high-risk resectable melanoma: A prospective phase II trial
Abstract
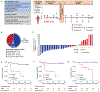
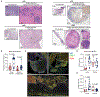
Intratumoral TLR9 agonists and anti-PD-1 produce clinical responses and broad immune activation. We conducted a single-arm study of neoadjuvant TLR9 agonist vidutolimod combined with anti-PD-1 nivolumab in high-risk resectable melanoma. In 31 evaluable patients, 55% major pathologic response (MPR) was observed, meeting primary endpoint. MPR was associated with necrosis, and melanophagocytosis with increased CD8+ tumor-infiltrating lymphocytes and plasmacytoid dendritic cells (pDCs) in the tumor microenvironment, and increased frequencies of Ki67+CD8+ T cells peripherally. MPRs had an enriched pre-treatment gene signature of myeloid cells, and response to therapy was associated with gene signatures of immune cells, pDCs, phagocytosis, and macrophage activation. MPRs gut microbiota were enriched for Gram-negative bacteria belonging to the Bacteroidaceae and Enterobacteriaceae families and the small subgroup of Gram-negative Firmicutes. Our findings support that combined vidutolimod and nivolumab stimulates a broad anti-tumor immune response and is associated with distinct baseline myeloid gene signature and gut microbiota. ClinicalTrials.gov identifier: NCT03618641.
Keywords: ICI; PD-1; TLR9; immunotherapy; innate agonist; macrophage; melanoma; neoadjuvant; pDC; vidutolimod.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.D. reports grants/research support (NIH/NCI and Checkmate Pharmaceuticals) and consulting (Checkmate Pharmaceuticals) during the conduct of the study. D.D. also reports grants/research support (Arcus, Immunocore, Merck, Regeneron Pharmaceuticals Inc., Tesaro/GSK.), consulting (ACM Bio, Ascendis, Castle, Clinical Care Options [CCO], Gerson Lehrman Group [GLG], Immunitas, Medical Learning Group [MLG], Replimmune, Trisalus, Xilio Therapeutics), speakers’ bureau (Castle Biosciences), steering committee membership (Immunocore, Replimmune) and patents related to gut microbial signatures of response and toxicity to immune checkpoint blockade (US Patent 63/124,231 and US Patent 63/208,719) outside the submitted work. P.D. is currently employed by Nanostring Technologies and reports stock options. Y.G.N. reports grants/research support (Bristol-Myers Squibb, Merck Sharp & Dohme and Pfizer) and consulting (Checkmate Pharmaceuticals) outside the submitted work. J.J.L. reports grants/research support (multiple), membership on data safety monitoring boards (multiple), membership on scientific advisory boards with no stock ownership or stock options (multiple), membership on scientific advisory boards with stock for (multiple), consulting (multiple) and a provisional patent for cancer immunotherapy (PCT/US18/36052: Microbiome Biomarkers for anti-PD-1/PD-L1 responsiveness: diagnostic, prognostic and therapeutic uses thereof) all outside the submitted work. D.M. is currently employed by Codiak Biosciences and reports stock options. D.B., J.W. and A.K. were formerly employed by CheckMate and report stock options. M.G.F. is currently employed by Regeneron Pharmaceuticals Inc. and reports stock options. J.M.K. reports grants/research support (Bristol-Myers Squibb, Amgen Inc.) and consulting (Bristol-Myers Squibb, Checkmate Pharmaceuticals, Novartis, Amgen Inc., Checkmate, Castle Biosciences, Inc., Immunocore LLC, Iovance, Novartis.) outside the submitted work. J.M.T. reports grants and consulting from Bristol-Myers Squibb, Merck Sharp & Dohme, Astra Zeneca, and Compugen outside the submitted work. H.M.Z. reports grants/research support (NIH/NCI and Checkmate Pharmaceuticals) and consulting (Checkmate Pharmaceuticals) during the conduct of the study. H.M.Z. also reports grants/research support (NIH/NCI, Bristol-Myers Squibb and GlaxoSmithKline), personal fees (GlaxoSmithKline, Bayer, and Vedanta) and pending provisional patents related to gut microbial signatures of response and toxicity to immune checkpoint blockade (US Patent 63/124,231 and US Patent 63/208,719) outside the submitted work.
Figures






References
-
- Eggermont AMM, Blank CU, Mandala M, Long GV, Atkinson VG, Dalle S, Haydon AM, Meshcheryakov A, Khattak A, Carlino MS, et al. (2021). Adjuvant pembrolizumab versus placebo in resected stage III melanoma (EORTC 1325-MG/KEYNOTE-054): distant metastasis-free survival results from a double-blind, randomised, controlled, phase 3 trial. Lancet Oncol 22, 643–654. 10.1016/S1470-2045(21)00065-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

