Degradation of IKZF1 prevents epigenetic progression of T cell exhaustion in an antigen-specific assay
- PMID: 39486420
- PMCID: PMC11604474
- DOI: 10.1016/j.xcrm.2024.101804
Degradation of IKZF1 prevents epigenetic progression of T cell exhaustion in an antigen-specific assay
Abstract
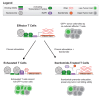
In cancer, chronic antigen stimulation drives effector T cells to exhaustion, limiting the efficacy of T cell therapies. Recent studies have demonstrated that epigenetic rewiring governs the transition of T cells from effector to exhausted states and makes a subset of exhausted T cells non-responsive to PD1 checkpoint blockade. Here, we describe an antigen-specific assay for T cell exhaustion that generates T cells phenotypically and transcriptionally similar to those found in human tumors. We perform a screen of human epigenetic regulators, identifying IKZF1 as a driver of T cell exhaustion. We determine that the IKZF1 degrader iberdomide prevents exhaustion by blocking chromatin remodeling at T cell effector enhancers and preserving the binding of AP-1, NF-κB, and NFAT. Thus, our study uncovers a role for IKZF1 as a driver of T cell exhaustion through epigenetic modulation, providing a rationale for the use of iberdomide in solid tumors to prevent T cell exhaustion.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests J.D.B. holds patents related to ATAC-seq and is an SAB member of Camp4 and seqWell. G.B., E.J., A.G., I.K.R., A.S.D., L.P., J.K., M.G.O., S.C., and D.A.M. are current or former employees of AstraZeneca. V.B.-W. is an employee of Revvity. S.L.T. was formerly an employee of Revvity.
Figures




References
-
- Friedrich M.J., Neri P., Kehl N., Michel J., Steiger S., Kilian M., Leblay N., Maity R., Sankowski R., Lee H., et al. The pre-existing T cell landscape determines the response to bispecific T cell engagers in multiple myeloma patients. Cancer Cell. 2023;41:711–725.e6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

