Pharmacological activation of STAT1-GSDME pyroptotic circuitry reinforces epigenetic immunotherapy for hepatocellular carcinoma
- PMID: 39486886
- PMCID: PMC12013592
- DOI: 10.1136/gutjnl-2024-332281
Pharmacological activation of STAT1-GSDME pyroptotic circuitry reinforces epigenetic immunotherapy for hepatocellular carcinoma
Abstract
Background: Genomic screening uncovered interferon-gamma (IFNγ) pathway defects in tumours refractory to immune checkpoint blockade (ICB). However, its non-mutational regulation and reversibility for therapeutic development remain less understood.
Objective: We aimed to identify ICB resistance-associated druggable histone deacetylases (HDACs) and develop a readily translatable combination approach for patients with hepatocellular carcinoma (HCC).
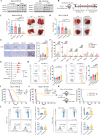
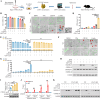
Design: We correlated the prognostic outcomes of HCC patients from a pembrolizumab trial (NCT03419481) with tumourous cell expressions of all HDAC isoforms by single-cell RNA sequencing. We investigated the therapeutic efficacy and mechanism of action of selective HDAC inhibition in 4 ICB-resistant orthotopic and spontaneous models using immune profiling, single-cell multiomics and chromatin immunoprecipitation-sequencing and verified by genetic modulations and co-culture systems.
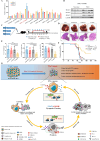
Results: HCC patients showing higher HDAC1/2/3 expressions exhibited deficient IFNγ signalling and poorer survival on ICB therapy. Transient treatment of a selective class-I HDAC inhibitor CXD101 resensitised HDAC1/2/3high tumours to ICB therapies, resulting in CD8+T cell-dependent antitumour and memory T cell responses. Mechanistically, CXD101 synergised with ICB to stimulate STAT1-driven antitumour immunity through enhanced chromatin accessibility and H3K27 hyperacetylation of IFNγ-responsive genes. Intratumoural recruitment of IFNγ+GZMB+cytotoxic lymphocytes further promoted cleavage of CXD101-induced Gasdermin E (GSDME) to trigger pyroptosis in a STAT1-dependent manner. Notably, deletion of GSDME mimicked STAT1 knockout in abolishing the antitumour efficacy and survival benefit of CXD101-ICB combination therapy by thwarting both pyroptotic and IFNγ responses.
Conclusion: Our immunoepigenetic strategy harnesses IFNγ-mediated network to augment the cancer-immunity cycle, revealing a self-reinforcing STAT1-GSDME pyroptotic circuitry as the mechanistic basis for an ongoing phase-II trial to tackle ICB resistance (NCT05873244).
Keywords: HEPATOCELLULAR CARCINOMA; IMMUNOTHERAPY; INTERFERON.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: The authors declare no conflicts of interest related to this work, except for the following declarations. SLC serves as an advisory member for AstraZeneca, MSD, Eisai, BMS, Ipsen and Hengrui, received research funds from MSD, Eisai, Ipsen, SIRTEX and Zailab, and honoraria from AstraZeneca, Eisai, Roche, Ipsen and MSD.
Figures





References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
