Spatiotemporal coordination of actin regulators generates invasive protrusions in cell-cell fusion
- PMID: 39487253
- PMCID: PMC12164543
- DOI: 10.1038/s41556-024-01541-5
Spatiotemporal coordination of actin regulators generates invasive protrusions in cell-cell fusion
Abstract
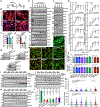
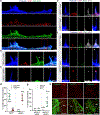
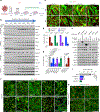
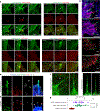
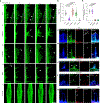
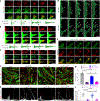
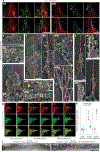
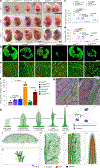
Invasive membrane protrusions play a central role in a variety of cellular processes. Unlike filopodia, invasive protrusions are mechanically stiff and propelled by branched actin polymerization. However, how branched actin filaments are organized to create finger-like invasive protrusions is unclear. Here, by examining the mammalian fusogenic synapse, where invasive protrusions are generated to promote cell membrane juxtaposition and fusion, we have uncovered the mechanism underlying invasive protrusion formation. We show that two nucleation-promoting factors for the Arp2/3 complex, WAVE and N-WASP, exhibit different localization patterns in the protrusions. Whereas WAVE is closely associated with the plasma membrane at the leading edge of the protrusive structures, N-WASP is enriched with WIP along the actin bundles in the shafts of the protrusions. During protrusion initiation and growth, the Arp2/3 complex nucleates branched actin filaments to generate low-density actin clouds in which the large GTPase dynamin organizes the new branched actin filaments into bundles, followed by actin-bundle stabilization by WIP, the latter functioning as an actin-bundling protein. Disruption of any of these components results in defective protrusions and failed myoblast fusion in cultured cells and mouse embryos. Together, our study has revealed the intricate spatiotemporal coordination between two nucleation-promoting factors and two actin-bundling proteins in building invasive protrusions at the mammalian fusogenic synapse and has general implications in understanding invasive protrusion formation in cellular processes beyond cell-cell fusion.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures








References
MeSH terms
Substances
Grants and funding
- R35GM136316/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 AR052777/AR/NIAMS NIH HHS/United States
- R35 GM140832/GM/NIGMS NIH HHS/United States
- R01AR052777/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R35 GM136316/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

