Biofilm formation and antibiogram profile of bacteria from infected wounds in a general hospital in southern Ethiopia
- PMID: 39487302
- PMCID: PMC11530625
- DOI: 10.1038/s41598-024-78283-9
Biofilm formation and antibiogram profile of bacteria from infected wounds in a general hospital in southern Ethiopia
Abstract
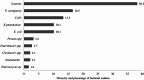
Biofilm-producing bacteria associated with wound infections exhibit exceptional drug resistance, leading to an escalation in morbidity, worse clinical outcomes (including delay in the healing process), and an increase in health care cost, burdening the whole system. This study is an attempt to estimate the prevalence and the relationship between the biofilm-forming capacity and multi-drug resistance of wound bacterial isolates. The findings intended to help clinicians, healthcare providers and program planners and to formulate an evidence-based decision-making process, especially in resource-limited healthcare settings. This study was done to assess the prevalence of bacterial infections in wounds and the antibiogram and biofilm-forming capacity of those bacteria in patients with clinical signs and symptoms, attending a General Hospital in southern Ethiopia. A cross-sectional study was performed in Arba Minch General Hospital from June to November 2021. The study participants comprised 201 patients with clinically infected wounds. Demographic and clinical data were gathered via a structured questionnaire. Specimens from wounds were taken from each participant and inoculated onto a series of culture media, namely MacConkey agar, mannitol salt agar, and blood agar, and different species were identified using a number of biochemical tests. Antimicrobial susceptibility tests were performed by means of the Kirby-Bauer disc diffusion technique following the guidelines of the Clinical and Laboratory Standards Institute. A micro-titer plate method was employed to detect the extent of biofilm formation. Bivariable and multivariable logistic regression models were applied to analyse the association between dependent and independent variables, and P values ≤ 0.05 were considered as statistically significant. Data analyses were done with Statistical Package for the Social Sciences version 25. Out of the 201 clinically infected wounds, 165 were found culture-positive with an overall prevalence of 82% (95% CI: 75.9-86.9). In total, 188 bacteria were recovered; 53.1% of them were Gram-positive cocci. The often-isolated bacterial species were Staphylococcus aureus, 38.3% (n = 72), and Pseudomonas aeruginosa, 16.4% (n = 31). The Gram-positive isolates showed considerable resistance against penicillin, 70%, and somewhat strong resistance against tetracycline, 57.7%. Gram-negative isolates showed severe resistance to ampicillin, 80.68%. The overall multi-drug resistance (MDR) among isolates was 48.4%. Extended beta-lactamase (ESBL)-producing Gram-negatives and methicillin-resistant Staphylococcus aureus (MRSA) accounted for 49 and 41.67%, respectively; 62.2% of the isolates were biofilm formers and were correlated statistically with MDR, ESBL producers, and MRSA (P < 0.005). The extent of biofilm formation and the prevalence of MDR bacteria associated with infected wounds hint at a public health threat that needs immediate attention. Thus, a more balanced and comprehensive wound management approach and antimicrobial stewardship program are essential in the study setting.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Swanson, T. et al. IWII Wound Infection in clinical practice consensus document: 2022 update. J. Wound Care. 31 (Sup12), S10–S21. 10.12968/jowc.2022.31.Sup12.S10 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases