Combined transcriptome and proteome profiling reveal cell-type-specific functions of Drosophila garland and pericardial nephrocytes
- PMID: 39487357
- PMCID: PMC11530456
- DOI: 10.1038/s42003-024-07062-z
Combined transcriptome and proteome profiling reveal cell-type-specific functions of Drosophila garland and pericardial nephrocytes
Abstract
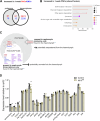
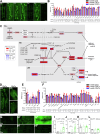
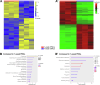
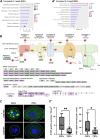
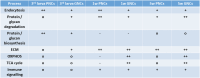
Drosophila nephrocytes are specialised cells that share critical functional, morphological, and molecular features with mammalian podocytes. Accordingly, nephrocytes represent a preferred invertebrate model for human glomerular disease. Here, we established a method for cell-specific isolation of the two types of Drosophila nephrocytes, garland and pericardial cells, from animals of different developmental stages and ages. Mass spectrometry-based proteomics and RNA-Seq-based transcriptomics were applied to characterise the proteome and transcriptome of the respective cells in an integrated and complementary manner. We observed characteristic changes in the proteome and transcriptome due to cellular ageing. Furthermore, functional enrichment analyses suggested that larval and adult nephrocytes, as well as garland and pericardial nephrocytes, fulfil distinct physiological functions. In addition, the pericardial nephrocytes were characterised by transcriptomic and proteomic profiles suggesting an atypical energy metabolism with very low oxidative phosphorylation rates. Moreover, the nephrocytes displayed typical signatures of extensive immune signalling and showed an active antimicrobial response to an infection. Factor-specific comparisons identified novel candidate proteins either expressed and secreted by the nephrocytes or sequestered by them. The data generated in this study represent a valuable basis for a more specific application of the Drosophila model in analysing renal cell function in health and disease.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- PA 517/12-1; PA 517/12-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 944, TP7; SFB 1557, TP12/Deutsche Forschungsgemeinschaft (German Research Foundation)
- HA 6421/4-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 944, TP21/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1182, TP2/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

