STAG2 loss in Ewing sarcoma alters enhancer-promoter contacts dependent and independent of EWS::FLI1
- PMID: 39487368
- PMCID: PMC11624272
- DOI: 10.1038/s44319-024-00303-6
STAG2 loss in Ewing sarcoma alters enhancer-promoter contacts dependent and independent of EWS::FLI1
Abstract
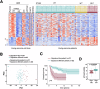
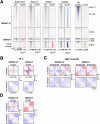
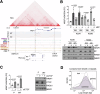
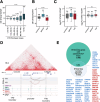
Cohesin complexes carrying STAG1 or STAG2 organize the genome into chromatin loops. STAG2 loss-of-function mutations promote metastasis in Ewing sarcoma, a pediatric cancer driven by the fusion transcription factor EWS::FLI1. We integrated transcriptomic data from patients and cellular models to identify a STAG2-dependent gene signature associated with worse prognosis. Subsequent genomic profiling and high-resolution chromatin interaction data from Capture Hi-C indicated that cohesin-STAG2 facilitates communication between EWS::FLI1-bound long GGAA repeats, presumably acting as neoenhancers, and their target promoters. Changes in CTCF-dependent chromatin contacts involving signature genes, unrelated to EWS::FLI1 binding, were also identified. STAG1 is unable to compensate for STAG2 loss and chromatin-bound cohesin is severely decreased, while levels of the processivity factor NIPBL remain unchanged, likely affecting DNA looping dynamics. These results illuminate how STAG2 loss modifies the chromatin interactome of Ewing sarcoma cells and provide a list of potential biomarkers and therapeutic targets.
Keywords: Cohesin; Genome Organization; Pediatric Cancer; Transcription.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures





References
-
- Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, Lareau CA, Ross L, Lin S, Dela Cruz FS et al (2021) Gene Expression Omnibus GSE1165977 (Hi-ChIP) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165977) [DATASET]
-
- Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, Lareau CA, Ross L, Lin S, Dela Cruz FS et al (2021) Gene Expression Omnibus GSE116495 (ChIP-seq) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116495) [DATASET]
MeSH terms
Substances
Grants and funding
- PROYE20046LOSA/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- PID2022-139333NB-I00/MCIN/AEI/10.13039/501100011033
- PID2019-106499RB-I00/MCIN/AEI/10.13039/501100011033
- PEJ-2020-AI/BMD-18406/Comunidad de Madrid (Community of Madrid)
- PRE2021-100005/MEC | Agencia Estatal de Investigación (AEI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

