Optimized psilocybin production in tryptophan catabolism-repressed fungi
- PMID: 39487767
- PMCID: PMC11530996
- DOI: 10.1111/1751-7915.70039
Optimized psilocybin production in tryptophan catabolism-repressed fungi
Abstract
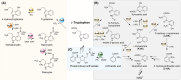
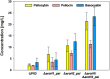
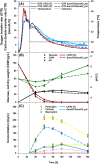
The high therapeutic potential of psilocybin, a prodrug of the psychotropic psilocin, holds great promise for the treatment of mental disorders such as therapy-refractory depression, alcohol use disorder and anorexia nervosa. Psilocybin has been designated a 'Breakthrough Therapy' by the US Food and Drug Administration, and therefore a sustainable production process must be established to meet future market demands. Here, we present the development of an in vivo psilocybin production chassis based on repression of l-tryptophan catabolism. We demonstrate the proof of principle in Saccharomyces cerevisiae expressing the psilocybin biosynthetic genes. Deletion of the two aminotransferase genes ARO8/9 and the indoleamine 2,3-dioxygenase gene BNA2 yielded a fivefold increase of psilocybin titre. We transferred this knowledge to the filamentous fungus Aspergillus nidulans and identified functional ARO8/9 orthologs involved in fungal l-tryptophan catabolism by genome mining and cross-complementation. The double deletion mutant of A. nidulans resulted in a 10-fold increased psilocybin production. Process optimization based on respiratory activity measurements led to a final psilocybin titre of 267 mg/L in batch cultures with a space-time-yield of 3.7 mg/L/h. These results demonstrate the suitability of our engineered A. nidulans to serve as a production strain for psilocybin and other tryptamine-derived pharmaceuticals.
© 2024 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Adams, A.M. , Kaplan, N.A. , Wei, Z. , Brinton, J.D. , Monnier, C.S. , Enacopol, A.L. et al. (2019) In vivo production of psilocybin in E. coli . Metabolic Engineering, 56, 111–119. - PubMed
-
- Anderlei, T. & Büchs, J. (2001) Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochemical Engineering Journal, 7, 157–162. - PubMed
-
- Blei, F. , Baldeweg, F. , Fricke, J. & Hoffmeister, D. (2018) Biocatalytic production of psilocybin and derivatives in tryptophan synthase‐enhanced reactions. Chemistry—a European Journal, 24, 10028–10031. - PubMed
-
- Bok, J.W. , Hoffmeister, D. , Maggio‐Hall, L.A. , Murillo, R. , Glasner, J.D. & Keller, N.P. (2006) Genomic mining for aspergillus natural products. Chemistry & Biology, 13, 31–37. - PubMed
-
- Bouhired, S. , Weber, M. , Kempf‐Sontag, A. , Keller, N.P. & Hoffmeister, D. (2007) Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genetics and Biology, 44, 1134–1145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

