Host-dependent specialized metabolism of nitrogen export in actinorhizal nodules induced by diazotrophic Actinomycetota Frankia cluster-2
- PMID: 39487991
- PMCID: PMC11850969
- DOI: 10.1093/jxb/erae446
Host-dependent specialized metabolism of nitrogen export in actinorhizal nodules induced by diazotrophic Actinomycetota Frankia cluster-2
Abstract
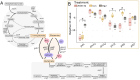
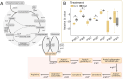
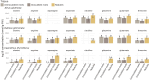
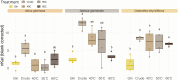
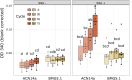
Frankia cluster-2 strains are diazotrophs that engage in root nodule symbiosis with actinorhizal plants of the Cucurbitales and the Rosales. Previous studies have shown that an assimilated nitrogen source, presumably arginine, is exported to the host in nodules of Datisca glomerata (Cucurbitales), while a different metabolite is exported in the nodules of Ceanothus thyrsiflorus (Rosales). To investigate if an assimilated nitrogen form is commonly exported to the host by cluster-2 strains, and which metabolite would be exported in Ceanothus, we analysed gene expression levels, metabolite profiles, and enzyme activities in nodules. We conclude that the export of assimilated nitrogen in symbiosis seems to be a common feature for Frankia cluster-2 strains, but the source of nitrogen is host dependent. The export of assimilated ammonium to the host suggests that 2-oxoglutarate is drawn from the tricarboxylic acid (TCA) cycle at a high rate. This specialized metabolism obviates the need for the reductive branch of the TCA cycle. We found that several genes encoding enzymes of central carbon and nitrogen metabolism were lacking in Frankia cluster-2 genomes: the glyoxylate shunt and succinate semialdehyde dehydrogenase. This led to a linearization of the TCA cycle, and we hypothesized that this could explain the low saprotrophic potential of Frankia cluster-2.
Keywords: Frankia; Actinorhizal symbiosis; GS synthetase; TCA cycle; nitrogenase; root nodules; succinic semialdehyde dehydrogenase.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Comparative Analysis of the Nodule Transcriptomes of Ceanothus thyrsiflorus (Rhamnaceae, Rosales) and Datisca glomerata (Datiscaceae, Cucurbitales).Front Plant Sci. 2018 Nov 14;9:1629. doi: 10.3389/fpls.2018.01629. eCollection 2018. Front Plant Sci. 2018. PMID: 30487804 Free PMC article.
-
Genome sequence of "Candidatus Frankia datiscae" Dg1, the uncultured microsymbiont from nitrogen-fixing root nodules of the dicot Datisca glomerata.J Bacteriol. 2011 Dec;193(24):7017-8. doi: 10.1128/JB.06208-11. J Bacteriol. 2011. PMID: 22123767 Free PMC article.
-
The Influence of the Host Plant Is the Major Ecological Determinant of the Presence of Nitrogen-Fixing Root Nodule Symbiont Cluster II Frankia Species in Soil.Appl Environ Microbiol. 2016 Dec 15;83(1):e02661-16. doi: 10.1128/AEM.02661-16. Print 2017 Jan 1. Appl Environ Microbiol. 2016. PMID: 27795313 Free PMC article.
-
Symbiosis between Frankia and actinorhizal plants: root nodules of non-legumes.Indian J Exp Biol. 2003 Oct;41(10):1165-83. Indian J Exp Biol. 2003. PMID: 15242283 Review.
-
Paradigms of actinorhizal symbiosis under the regime of global climatic changes: New insights and perspectives.J Basic Microbiol. 2022 Jul;62(7):764-778. doi: 10.1002/jobm.202200043. Epub 2022 May 31. J Basic Microbiol. 2022. PMID: 35638879 Review.
References
-
- Alloisio N, Queiroux C, Fournier P, Pujic P, Normand P, Vallenet D, Médigue C, Yamaura M, Kakoi K, Kucho K.. 2010. The Frankia alni symbiotic transcriptome. Molecular Plant-Microbe Interactions 23, 593–607. - PubMed
-
- Behrmann I, Hillemann D, Pühler A, Strauch E, Wohlleben W.. 1990. Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. Journal of Bacteriology 172, 5326–5334. - PMC - PubMed

