Single-molecule imaging and molecular dynamics simulations reveal early activation of the MET receptor in cells
- PMID: 39488533
- PMCID: PMC11531568
- DOI: 10.1038/s41467-024-53772-7
Single-molecule imaging and molecular dynamics simulations reveal early activation of the MET receptor in cells
Abstract
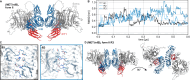
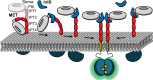
Embedding of cell-surface receptors into a membrane defines their dynamics but also complicates experimental characterization of their signaling complexes. The hepatocyte growth factor receptor MET is a receptor tyrosine kinase involved in cellular processes such as proliferation, migration, and survival. It is also targeted by the pathogen Listeria monocytogenes, whose invasion protein, internalin B (InlB), binds to MET, forming a signaling dimer that triggers pathogen internalization. Here we use an integrative structural biology approach, combining molecular dynamics simulations and single-molecule Förster resonance energy transfer (smFRET) in cells, to investigate the early stages of MET activation. Our simulations show that InlB binding stabilizes MET in a conformation that promotes dimer formation. smFRET reveals that the in situ dimer structure closely resembles one of two previously published crystal structures, though with key differences. This study refines our understanding of MET activation and provides a methodological framework for studying other plasma membrane receptors.
© 2024. The Author(s).
Conflict of interest statement
L.I. and H.d.J are listed as co-founders of the company Ardis S.r.l. All other authors declare no competing interests.
Figures

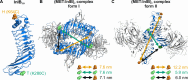


References
Publication types
MeSH terms
Substances
Grants and funding
- CRC 1507/Deutsche Forschungsgemeinschaft (German Research Foundation)
- INST 161/778-1 FUGG/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRC 1507/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRC 1507/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRC 1507/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

