Five-gene prognostic model based on autophagy-dependent cell death for predicting prognosis in lung adenocarcinoma
- PMID: 39488588
- PMCID: PMC11531468
- DOI: 10.1038/s41598-024-76186-3
Five-gene prognostic model based on autophagy-dependent cell death for predicting prognosis in lung adenocarcinoma
Abstract
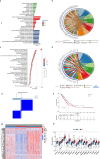
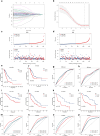
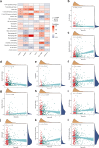
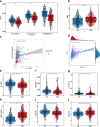
Non-small cell lung adenocarcinoma (LUAD) is the predominant form of lung cancer originating from lung epithelial cells, making it the most prevalent pathological type. Currently, reliable indicators for predicting treatment efficacy and disease prognosis are lacking. Despite extensive validation of autophagy-dependent cell death (ADCD) in solid tumor studies and its correlation with immunotherapy effectiveness and cancer prognosis, systematic research on ADCD-related genes in LUAD is limited. We utilized AddModuleScore, ssGSEA, and WGCNA to identify genes associated with ADCD across single-cell and bulk transcriptome datasets. The TCGA dataset, comprising 598 cases, was randomly divided into training and validation sets to develop an ADCD-related LUAD prediction model. Internal validation was performed using the TCGA validation set. For external validation, datasets GSE13213 (119 LUAD samples), GSE26939 (115 LUAD samples), GSE29016 (39 LUAD samples), and GSE30219 (86 LUAD samples) were employed. We evaluated the model's accuracy and effectiveness in predicting prognostic risk. Additionally, CIBERSORT, ESTIMATE, and ssGSEA techniques were used to explore immunological characteristics, drug response, and gene expression in LUAD. Real-time RT-PCR was conducted to assess variations in mRNA expression levels of the gene XCR1 between cancerous and normal tissues in 10 lung cancer patients. We identified 249 genes associated with autophagy-dependent cell death (ADCD) at both single-cell and bulk transcriptome levels. Univariate COX regression analysis revealed that 18 genes were significantly associated with overall survival (OS). Using LASSO-Cox analysis, we developed an ADCD signature based on five genes (BIRC3, TAP1, SLAMF1, XCR1, and HLA-DMB) and created the ADCD-related risk scoring system (ADCDRS). Validation of this model demonstrated its ability to predict disease prognosis and its correlation with clinical characteristics, immune cell infiltration, and the tumor microenvironment. To enhance clinical applicability, we integrated an ADCDRS nomogram. Furthermore, we identified potential drugs targeting specific risk subgroups. We successfully identified a model based on five ADCD genes to predict disease prognosis and treatment efficacy in LUAD, as well as to assess the tumor immune microenvironment. An efficient and practical ADCDRS nomogram was designed.
Keywords: Autophagy-dependent cell death; Immunotherapy; Lung adenocarcinoma; Non-small cell lung cancer; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

