Genomic and transcriptomic landscape of human gastrointestinal stromal tumors
- PMID: 39489749
- PMCID: PMC11532483
- DOI: 10.1038/s41467-024-53821-1
Genomic and transcriptomic landscape of human gastrointestinal stromal tumors
Abstract
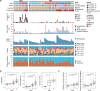
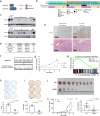
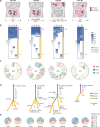
Gastrointestinal stromal tumor (GISTs) are clinically heterogenous exhibiting varying degrees of disease aggressiveness in individual patients. We comprehensively describe the genomic and transcriptomic landscape of a cohort of 117 GISTs including 31 low-risk, 18 intermediate-risk, 29 high-risk, 34 metastatic and 5 neoadjuvant GISTs from 105 patients. GISTs have notably low tumor mutation burden but widespread copy number variations. Aggressive GISTs harbor remarkably more genomic aberrations than low-/intermediate-risk GISTs. Complex genomic alterations, chromothripsis and kataegis, occur selectively in aggressive GISTs. Despite the paucity of mutations, recurrent inactivating YLPM1 mutations are identified (10.3%, 7 of 68 patients), enriched in high-risk/metastatic GIST and functional study further demonstrates YLPM1 inactivation promotes GIST proliferation, growth and oxidative phosphorylation. Spatially and temporally separated GISTs from individual patients demonstrate complex tumor heterogeneity in metastatic GISTs. Finally, four prominent subtypes are proposed with different genomic features, expression profiles, immune characteristics, clinical characteristics and subtype-specific treatment strategies. This large-scale analysis depicts the landscape and provides further insights into GIST pathogenesis and precise treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hirota, S. et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science279, 577–580 (1998). - PubMed
-
- Heinrich, M. C. et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science299, 708–710 (2003). - PubMed
-
- Corless, C. L., Barnett, C. M. & Heinrich, M. C. Gastrointestinal stromal tumours: origin and molecular oncology. Nat. Rev. Cancer11, 865–878 (2011). - PubMed
-
- Blay, J. Y., Kang, Y. K., Nishida, T. & von Mehren, M. Gastrointestinal stromal tumours. Nat. Rev. Dis. Prim.7, 22 (2021). - PubMed
-
- Abraham, S. C., Krasinskas, A. M., Hofstetter, W. L., Swisher, S. G. & Wu, T. T. Seedling” mesenchymal tumors are common incidental tumors of the esophagogastric junction. Am. J. Surg. Pathol.31, 1629–1635 (2007). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

