N6-Methyladenosine modification activates the serine synthesis pathway to mediate therapeutic resistance in liver cancer
- PMID: 39489921
- PMCID: PMC11638877
- DOI: 10.1016/j.ymthe.2024.10.025
N6-Methyladenosine modification activates the serine synthesis pathway to mediate therapeutic resistance in liver cancer
Abstract
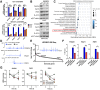
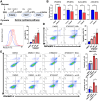
Metabolic adaptation serves as a significant driving force for cancer growth and poses a substantial obstacle for cancer therapies. Herein, we unraveled the role of m6A-mediated serine synthesis pathway (SSP) regulation in both hepatocellular carcinoma (HCC) development and therapeutic resistance. We demonstrated that treatment of highly specific m6A inhibitor (STM2457) effectively inhibited HCC cell line growth and suppressed spontaneous HCC formation in mice driven by liver-specific Tp53 knockout and Myc overexpression. Using GLORI-seq, we delineated a single-base-resolution m6A landscape in human HCC cell lines. Interestingly, we identified three core enzymes in the SSP (PHGDH, PSAT1, and PSPH) as novel targets of METTL3-mediated m6A modification. In these SSP genes, m6A modification recruited m6A reader IGF2BP3 to stabilize their mRNA transcripts, thereby enhancing their mRNA and protein expression in HCC cells. Most importantly, our GLORI-seq data revealed that sorafenib-resistant HCC cells elevated m6A modification in SSP genes to promote protein expression and antioxidant production. STM2457 treatment attenuated the serine synthesis pathway, induced oxidative stress, and sensitized HCC cells to sorafenib and lenvatinib treatments. In conclusion, our findings suggest that targeting m6A could be a potential therapeutic strategy for HCC treatment.
Keywords: IGF2BP3; METTL3; SSP pathway; STM2457; TKI treatment; liver cancer; m6A; m6A modification.
Copyright © 2024 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

