Equine ANP32 proteins support influenza A virus RNA polymerase activity
- PMID: 39491182
- PMCID: PMC10786659
- DOI: 10.1016/j.virs.2023.10.009
Equine ANP32 proteins support influenza A virus RNA polymerase activity
Abstract
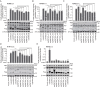
Host ANP32 family proteins are crucial for maintaining the activity of influenza RNA polymerase and play an important role in the cross-species transmission of influenza viruses. To date, the molecular properties of equine ANP32 (eqANP32) protein are poorly understood, particularly the mechanisms that affect equine influenza virus (EIV) RNA polymerase activity. Here, we found that there are six alternative splicing variants of equine ANP32A (eqANP32A) with different levels of expression. Further studies showed that these six splicing variants of eqANP32A supported the activity of EIV RNA polymerase to varying degrees, with the variant eqANP32A_X2 having the highest expression abundance and exhibiting the highest support of polymerase activity. Sequence analysis demonstrated that the differences in the N-Cap regions of the six splicing variants significantly affected their N-terminal conformation, but did not affect their ability to bind RNA polymerase. We also demonstrated that there is only one transcript of eqANP32B, and that this transcript showed only very low support to the EIV RNA polymerase. This functional defect in eqANP32B is caused by the sequence of the 110-259 amino acids at its C-terminus. Our results indicated that it is the eqANP32A_X2 protein that mainly determines the efficiency of the EIV replication in horses. In conclusion, our study parsed the molecular properties of eqANP32 family proteins and revealed the sequence features of eqANP32A and eqANP32B, suggesting for the first time that the N-cap region of ANP32A protein also plays an important role in supporting the activity of the influenza virus polymerase.
Keywords: Equine influenza virus (EIV); N-Cap domain; RNA polymerase activity; equine ANP32A; equine ANP32B.
Copyright © 2023 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflicts of interest, financial or otherwise.
Figures



References
-
- Bradel-Tretheway B.G., Mattiacio J.L., Krasnoselsky A., Stevenson C., Purdy D., Dewhurst S., Katze M.G. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and rna polymerase accessory factors. J. Virol. 2011;85:8569–8581. - PMC - PubMed
-
- Brister H., Barnum S.M., Reedy S., Chambers T.M., Pusterla N. Validation of two multiplex real-time pcr assays based on single nucleotide polymorphisms of the ha1 gene of equine influenza a virus in order to differentiate between clade 1 and clade 2 Florida sublineage isolates. J. Vet. Diagn. Invest. 2019;31:137–141. - PMC - PubMed
-
- Brody H. Influenza. Nature. 2019;573:S49. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

